
Solemya velesiana gill symbiont
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Gammaproteobacteria incertae sedis; sulfur-oxidizing symbionts
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
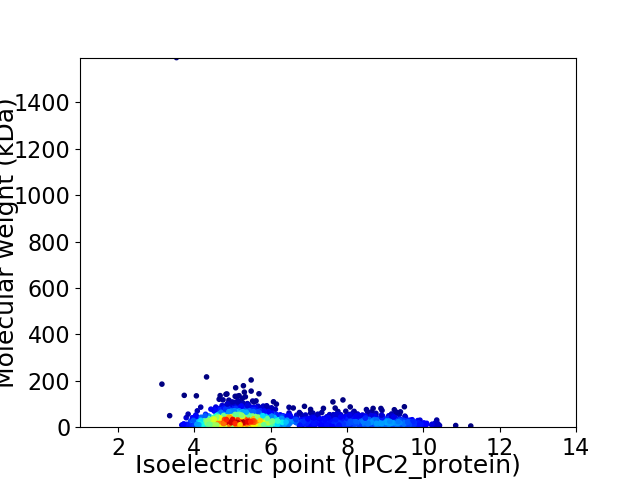
Virtual 2D-PAGE plot for 2304 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
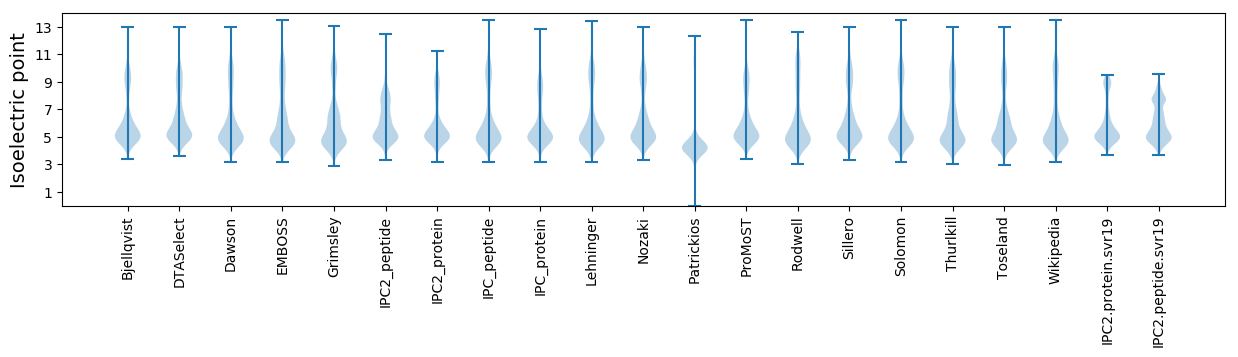
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1T2KSF8|A0A1T2KSF8_9GAMM Uncharacterized protein OS=Solemya velesiana gill symbiont OX=1918948 GN=BOW51_10330 PE=4 SV=1
MM1 pKa = 7.34NIKK4 pKa = 9.76VRR6 pKa = 11.84EE7 pKa = 3.97IMKK10 pKa = 9.76KK11 pKa = 10.31CGIVVCAALYY21 pKa = 10.13IATPAFADD29 pKa = 3.49DD30 pKa = 3.91VEE32 pKa = 5.01IFFNTSNVSGEE43 pKa = 4.6DD44 pKa = 2.94IGDD47 pKa = 3.93PLVMFGLDD55 pKa = 3.35YY56 pKa = 10.8RR57 pKa = 11.84PNVINANLCNAATLAEE73 pKa = 4.14VQNCGWDD80 pKa = 3.71DD81 pKa = 3.68PTTTDD86 pKa = 5.32DD87 pKa = 3.98DD88 pKa = 4.28DD89 pKa = 5.68FYY91 pKa = 11.76AAYY94 pKa = 10.98YY95 pKa = 10.75NVFTNEE101 pKa = 3.69DD102 pKa = 3.54VADD105 pKa = 4.51GSLSFLEE112 pKa = 4.08VLRR115 pKa = 11.84ASLRR119 pKa = 11.84YY120 pKa = 9.72VLGEE124 pKa = 4.07LDD126 pKa = 3.82DD127 pKa = 3.97VRR129 pKa = 11.84IGLMLNHH136 pKa = 6.3AQINNCEE143 pKa = 4.26GASGTAGCSNGGYY156 pKa = 9.52IAMGLLPIDD165 pKa = 4.4AVDD168 pKa = 5.09NSGGWYY174 pKa = 6.55TTWLDD179 pKa = 3.28NDD181 pKa = 3.79GDD183 pKa = 4.35GVHH186 pKa = 7.15DD187 pKa = 4.42VDD189 pKa = 6.02AGDD192 pKa = 5.15DD193 pKa = 3.78PSTGLNILDD202 pKa = 4.31GKK204 pKa = 10.01LASILDD210 pKa = 4.09LGTGAGAHH218 pKa = 5.84SNQGKK223 pKa = 8.86EE224 pKa = 4.4LYY226 pKa = 10.32FEE228 pKa = 4.87LYY230 pKa = 9.98RR231 pKa = 11.84YY232 pKa = 9.85LRR234 pKa = 11.84GWDD237 pKa = 3.11IYY239 pKa = 10.69NGHH242 pKa = 6.46VGYY245 pKa = 11.0ADD247 pKa = 3.48YY248 pKa = 11.14DD249 pKa = 3.95DD250 pKa = 6.02RR251 pKa = 11.84CDD253 pKa = 4.71EE254 pKa = 5.55DD255 pKa = 5.56NLDD258 pKa = 4.53DD259 pKa = 5.12DD260 pKa = 4.78TMPSCFDD267 pKa = 3.1RR268 pKa = 11.84RR269 pKa = 11.84GNPEE273 pKa = 4.95DD274 pKa = 5.59DD275 pKa = 4.52DD276 pKa = 5.5DD277 pKa = 4.77YY278 pKa = 11.84PAIAWDD284 pKa = 3.58ATTVGDD290 pKa = 3.81VGIGATAPDD299 pKa = 4.27YY300 pKa = 10.73PVEE303 pKa = 4.11EE304 pKa = 4.77AGNTEE309 pKa = 4.48YY310 pKa = 11.2YY311 pKa = 10.86SPFEE315 pKa = 4.42DD316 pKa = 4.19QSLICSGVYY325 pKa = 10.31AINFMFGVSNQDD337 pKa = 2.84NDD339 pKa = 3.6SDD341 pKa = 5.32NEE343 pKa = 4.1IEE345 pKa = 5.85DD346 pKa = 4.72DD347 pKa = 4.33DD348 pKa = 5.45AGQISAEE355 pKa = 4.07SAGMVEE361 pKa = 5.54LDD363 pKa = 3.59GSDD366 pKa = 4.57PIDD369 pKa = 3.15ISGNRR374 pKa = 11.84NEE376 pKa = 4.65FGTLIEE382 pKa = 4.03WLYY385 pKa = 10.44EE386 pKa = 4.2GSDD389 pKa = 3.54GSPRR393 pKa = 11.84DD394 pKa = 3.59LSDD397 pKa = 3.22LAGVQNVTSYY407 pKa = 11.42FMYY410 pKa = 10.24KK411 pKa = 10.69GNVQNTMNGYY421 pKa = 9.51AVSGGSGQAYY431 pKa = 8.74EE432 pKa = 4.61VLDD435 pKa = 4.31DD436 pKa = 4.41PKK438 pKa = 11.85AMVDD442 pKa = 3.83TITNIFKK449 pKa = 10.63EE450 pKa = 4.17ILRR453 pKa = 11.84QNTTFEE459 pKa = 4.31APAVTVNSYY468 pKa = 10.73NRR470 pKa = 11.84LTHH473 pKa = 6.79RR474 pKa = 11.84DD475 pKa = 3.57EE476 pKa = 5.45LFFALFSQEE485 pKa = 4.06EE486 pKa = 4.22HH487 pKa = 6.26QNWPGNLKK495 pKa = 10.17KK496 pKa = 10.91YY497 pKa = 10.52KK498 pKa = 10.16LGEE501 pKa = 3.91ITTEE505 pKa = 3.73VCTDD509 pKa = 3.72LNGDD513 pKa = 4.34GDD515 pKa = 4.07TTDD518 pKa = 3.69PGEE521 pKa = 4.53CVTEE525 pKa = 3.97PVQRR529 pKa = 11.84LVDD532 pKa = 3.73QNDD535 pKa = 3.41NEE537 pKa = 4.84AVNQSDD543 pKa = 4.33GVFKK547 pKa = 10.58ATTCSFWSNCSIDD560 pKa = 3.72RR561 pKa = 11.84DD562 pKa = 3.79NDD564 pKa = 3.25GTADD568 pKa = 3.69ADD570 pKa = 3.84GDD572 pKa = 3.93IVEE575 pKa = 4.49WGGAAEE581 pKa = 5.05EE582 pKa = 4.32ITLSRR587 pKa = 11.84TLYY590 pKa = 10.2TYY592 pKa = 10.6IGTDD596 pKa = 3.0RR597 pKa = 11.84YY598 pKa = 10.45AVHH601 pKa = 7.31EE602 pKa = 4.45DD603 pKa = 3.29TSQITHH609 pKa = 5.93EE610 pKa = 4.4MLNVSLDD617 pKa = 3.67EE618 pKa = 4.46TSVHH622 pKa = 6.82AGDD625 pKa = 5.88DD626 pKa = 3.56ICDD629 pKa = 3.4VDD631 pKa = 4.45GDD633 pKa = 3.97SDD635 pKa = 4.62GDD637 pKa = 4.06FEE639 pKa = 5.01CVDD642 pKa = 3.47MDD644 pKa = 4.56GDD646 pKa = 3.99GDD648 pKa = 4.72ADD650 pKa = 4.03STDD653 pKa = 3.3HH654 pKa = 7.73DD655 pKa = 4.85LLRR658 pKa = 11.84EE659 pKa = 4.22DD660 pKa = 4.84LLQIARR666 pKa = 11.84SYY668 pKa = 11.3DD669 pKa = 3.53PEE671 pKa = 4.33TGEE674 pKa = 4.26VLQNMGDD681 pKa = 4.24PIHH684 pKa = 6.84ARR686 pKa = 11.84PAVIEE691 pKa = 3.92YY692 pKa = 10.43DD693 pKa = 3.52ADD695 pKa = 3.85LSNQGACPDD704 pKa = 3.94LRR706 pKa = 11.84IAMTTNEE713 pKa = 4.4GQFHH717 pKa = 7.2LIDD720 pKa = 4.26AGGDD724 pKa = 3.63SGCTDD729 pKa = 2.95NDD731 pKa = 3.82ANVGGTEE738 pKa = 4.19VVSFMPEE745 pKa = 3.4EE746 pKa = 4.49LLPQLKK752 pKa = 9.35QVNGPITVEE761 pKa = 4.02GTGQTRR767 pKa = 11.84FNTYY771 pKa = 10.49GLDD774 pKa = 3.47GSPVVWRR781 pKa = 11.84YY782 pKa = 9.93DD783 pKa = 3.12ADD785 pKa = 4.68AGDD788 pKa = 3.62GTARR792 pKa = 11.84DD793 pKa = 3.71GVINYY798 pKa = 8.94GADD801 pKa = 3.33SDD803 pKa = 4.09EE804 pKa = 4.59DD805 pKa = 3.74FVYY808 pKa = 10.84LFLTQRR814 pKa = 11.84RR815 pKa = 11.84GGKK818 pKa = 9.85GIWALDD824 pKa = 3.34ITNPTSPEE832 pKa = 3.99LMWKK836 pKa = 9.39VWDD839 pKa = 4.14GDD841 pKa = 3.88LTSVNGTSAADD852 pKa = 3.57VYY854 pKa = 11.57SEE856 pKa = 5.77LGQTWSTPKK865 pKa = 9.25LTKK868 pKa = 10.35IKK870 pKa = 10.65DD871 pKa = 3.41GEE873 pKa = 4.08YY874 pKa = 10.77DD875 pKa = 3.09RR876 pKa = 11.84GVLVFGGGYY885 pKa = 10.73DD886 pKa = 3.71PDD888 pKa = 3.66QDD890 pKa = 3.8DD891 pKa = 5.14AYY893 pKa = 11.31SFSDD897 pKa = 3.36TQGRR901 pKa = 11.84AIYY904 pKa = 10.53VVDD907 pKa = 4.95AYY909 pKa = 9.81TGEE912 pKa = 4.28PLWTVSGEE920 pKa = 4.25AASTTPTQPSGGASGTMLIPEE941 pKa = 4.72MDD943 pKa = 3.33NSIPAEE949 pKa = 4.06VQPADD954 pKa = 3.9IDD956 pKa = 3.81SDD958 pKa = 4.41GYY960 pKa = 11.04LDD962 pKa = 4.94RR963 pKa = 11.84IYY965 pKa = 11.18AVDD968 pKa = 3.33VVGRR972 pKa = 11.84VFRR975 pKa = 11.84IDD977 pKa = 5.0FDD979 pKa = 3.67ILPNGDD985 pKa = 3.57STSTQMYY992 pKa = 8.52GGGMVAALNDD1002 pKa = 3.78SSCDD1006 pKa = 3.33EE1007 pKa = 4.24EE1008 pKa = 6.17APTDD1012 pKa = 4.01GVADD1016 pKa = 3.61ACHH1019 pKa = 6.02RR1020 pKa = 11.84RR1021 pKa = 11.84FYY1023 pKa = 10.94NKK1025 pKa = 9.32PDD1027 pKa = 3.26VAVMVGYY1034 pKa = 8.61PAAPYY1039 pKa = 9.18VQVAVGSGYY1048 pKa = 10.22RR1049 pKa = 11.84AWPASVSGIQDD1060 pKa = 3.31RR1061 pKa = 11.84FYY1063 pKa = 11.69VLFDD1067 pKa = 3.95EE1068 pKa = 5.37NVIDD1072 pKa = 4.33PLLSSVYY1079 pKa = 9.13GTGAGEE1085 pKa = 4.1QYY1087 pKa = 10.81HH1088 pKa = 5.12WTEE1091 pKa = 5.01DD1092 pKa = 3.24SDD1094 pKa = 4.56LANLTDD1100 pKa = 3.4IPGDD1104 pKa = 3.41VSDD1107 pKa = 4.3YY1108 pKa = 11.51SGAQTAAEE1116 pKa = 3.88TALGNKK1122 pKa = 8.96HH1123 pKa = 5.02GWYY1126 pKa = 10.35FNLEE1130 pKa = 4.14LTTDD1134 pKa = 3.15AGGTVTSHH1142 pKa = 6.36EE1143 pKa = 4.54KK1144 pKa = 10.62VLSDD1148 pKa = 3.17SVTFNGLILFTTYY1161 pKa = 9.86IRR1163 pKa = 11.84SSEE1166 pKa = 4.17DD1167 pKa = 3.2EE1168 pKa = 4.54ANVCEE1173 pKa = 4.29PSLGSGRR1180 pKa = 11.84FYY1182 pKa = 11.31AVNAFNGLPVGDD1194 pKa = 4.02INQGVFFTEE1203 pKa = 3.84DD1204 pKa = 2.94TARR1207 pKa = 11.84SDD1209 pKa = 3.27FSEE1212 pKa = 4.06ADD1214 pKa = 3.04RR1215 pKa = 11.84YY1216 pKa = 10.21MNLGRR1221 pKa = 11.84KK1222 pKa = 8.66GIPTTPQIILTDD1234 pKa = 3.56EE1235 pKa = 4.53GPRR1238 pKa = 11.84VIVTTEE1244 pKa = 4.69LIPEE1248 pKa = 4.43GLLDD1252 pKa = 4.61SDD1254 pKa = 5.07LFSKK1258 pKa = 10.21KK1259 pKa = 7.04WWLDD1263 pKa = 3.16AKK1265 pKa = 10.96
MM1 pKa = 7.34NIKK4 pKa = 9.76VRR6 pKa = 11.84EE7 pKa = 3.97IMKK10 pKa = 9.76KK11 pKa = 10.31CGIVVCAALYY21 pKa = 10.13IATPAFADD29 pKa = 3.49DD30 pKa = 3.91VEE32 pKa = 5.01IFFNTSNVSGEE43 pKa = 4.6DD44 pKa = 2.94IGDD47 pKa = 3.93PLVMFGLDD55 pKa = 3.35YY56 pKa = 10.8RR57 pKa = 11.84PNVINANLCNAATLAEE73 pKa = 4.14VQNCGWDD80 pKa = 3.71DD81 pKa = 3.68PTTTDD86 pKa = 5.32DD87 pKa = 3.98DD88 pKa = 4.28DD89 pKa = 5.68FYY91 pKa = 11.76AAYY94 pKa = 10.98YY95 pKa = 10.75NVFTNEE101 pKa = 3.69DD102 pKa = 3.54VADD105 pKa = 4.51GSLSFLEE112 pKa = 4.08VLRR115 pKa = 11.84ASLRR119 pKa = 11.84YY120 pKa = 9.72VLGEE124 pKa = 4.07LDD126 pKa = 3.82DD127 pKa = 3.97VRR129 pKa = 11.84IGLMLNHH136 pKa = 6.3AQINNCEE143 pKa = 4.26GASGTAGCSNGGYY156 pKa = 9.52IAMGLLPIDD165 pKa = 4.4AVDD168 pKa = 5.09NSGGWYY174 pKa = 6.55TTWLDD179 pKa = 3.28NDD181 pKa = 3.79GDD183 pKa = 4.35GVHH186 pKa = 7.15DD187 pKa = 4.42VDD189 pKa = 6.02AGDD192 pKa = 5.15DD193 pKa = 3.78PSTGLNILDD202 pKa = 4.31GKK204 pKa = 10.01LASILDD210 pKa = 4.09LGTGAGAHH218 pKa = 5.84SNQGKK223 pKa = 8.86EE224 pKa = 4.4LYY226 pKa = 10.32FEE228 pKa = 4.87LYY230 pKa = 9.98RR231 pKa = 11.84YY232 pKa = 9.85LRR234 pKa = 11.84GWDD237 pKa = 3.11IYY239 pKa = 10.69NGHH242 pKa = 6.46VGYY245 pKa = 11.0ADD247 pKa = 3.48YY248 pKa = 11.14DD249 pKa = 3.95DD250 pKa = 6.02RR251 pKa = 11.84CDD253 pKa = 4.71EE254 pKa = 5.55DD255 pKa = 5.56NLDD258 pKa = 4.53DD259 pKa = 5.12DD260 pKa = 4.78TMPSCFDD267 pKa = 3.1RR268 pKa = 11.84RR269 pKa = 11.84GNPEE273 pKa = 4.95DD274 pKa = 5.59DD275 pKa = 4.52DD276 pKa = 5.5DD277 pKa = 4.77YY278 pKa = 11.84PAIAWDD284 pKa = 3.58ATTVGDD290 pKa = 3.81VGIGATAPDD299 pKa = 4.27YY300 pKa = 10.73PVEE303 pKa = 4.11EE304 pKa = 4.77AGNTEE309 pKa = 4.48YY310 pKa = 11.2YY311 pKa = 10.86SPFEE315 pKa = 4.42DD316 pKa = 4.19QSLICSGVYY325 pKa = 10.31AINFMFGVSNQDD337 pKa = 2.84NDD339 pKa = 3.6SDD341 pKa = 5.32NEE343 pKa = 4.1IEE345 pKa = 5.85DD346 pKa = 4.72DD347 pKa = 4.33DD348 pKa = 5.45AGQISAEE355 pKa = 4.07SAGMVEE361 pKa = 5.54LDD363 pKa = 3.59GSDD366 pKa = 4.57PIDD369 pKa = 3.15ISGNRR374 pKa = 11.84NEE376 pKa = 4.65FGTLIEE382 pKa = 4.03WLYY385 pKa = 10.44EE386 pKa = 4.2GSDD389 pKa = 3.54GSPRR393 pKa = 11.84DD394 pKa = 3.59LSDD397 pKa = 3.22LAGVQNVTSYY407 pKa = 11.42FMYY410 pKa = 10.24KK411 pKa = 10.69GNVQNTMNGYY421 pKa = 9.51AVSGGSGQAYY431 pKa = 8.74EE432 pKa = 4.61VLDD435 pKa = 4.31DD436 pKa = 4.41PKK438 pKa = 11.85AMVDD442 pKa = 3.83TITNIFKK449 pKa = 10.63EE450 pKa = 4.17ILRR453 pKa = 11.84QNTTFEE459 pKa = 4.31APAVTVNSYY468 pKa = 10.73NRR470 pKa = 11.84LTHH473 pKa = 6.79RR474 pKa = 11.84DD475 pKa = 3.57EE476 pKa = 5.45LFFALFSQEE485 pKa = 4.06EE486 pKa = 4.22HH487 pKa = 6.26QNWPGNLKK495 pKa = 10.17KK496 pKa = 10.91YY497 pKa = 10.52KK498 pKa = 10.16LGEE501 pKa = 3.91ITTEE505 pKa = 3.73VCTDD509 pKa = 3.72LNGDD513 pKa = 4.34GDD515 pKa = 4.07TTDD518 pKa = 3.69PGEE521 pKa = 4.53CVTEE525 pKa = 3.97PVQRR529 pKa = 11.84LVDD532 pKa = 3.73QNDD535 pKa = 3.41NEE537 pKa = 4.84AVNQSDD543 pKa = 4.33GVFKK547 pKa = 10.58ATTCSFWSNCSIDD560 pKa = 3.72RR561 pKa = 11.84DD562 pKa = 3.79NDD564 pKa = 3.25GTADD568 pKa = 3.69ADD570 pKa = 3.84GDD572 pKa = 3.93IVEE575 pKa = 4.49WGGAAEE581 pKa = 5.05EE582 pKa = 4.32ITLSRR587 pKa = 11.84TLYY590 pKa = 10.2TYY592 pKa = 10.6IGTDD596 pKa = 3.0RR597 pKa = 11.84YY598 pKa = 10.45AVHH601 pKa = 7.31EE602 pKa = 4.45DD603 pKa = 3.29TSQITHH609 pKa = 5.93EE610 pKa = 4.4MLNVSLDD617 pKa = 3.67EE618 pKa = 4.46TSVHH622 pKa = 6.82AGDD625 pKa = 5.88DD626 pKa = 3.56ICDD629 pKa = 3.4VDD631 pKa = 4.45GDD633 pKa = 3.97SDD635 pKa = 4.62GDD637 pKa = 4.06FEE639 pKa = 5.01CVDD642 pKa = 3.47MDD644 pKa = 4.56GDD646 pKa = 3.99GDD648 pKa = 4.72ADD650 pKa = 4.03STDD653 pKa = 3.3HH654 pKa = 7.73DD655 pKa = 4.85LLRR658 pKa = 11.84EE659 pKa = 4.22DD660 pKa = 4.84LLQIARR666 pKa = 11.84SYY668 pKa = 11.3DD669 pKa = 3.53PEE671 pKa = 4.33TGEE674 pKa = 4.26VLQNMGDD681 pKa = 4.24PIHH684 pKa = 6.84ARR686 pKa = 11.84PAVIEE691 pKa = 3.92YY692 pKa = 10.43DD693 pKa = 3.52ADD695 pKa = 3.85LSNQGACPDD704 pKa = 3.94LRR706 pKa = 11.84IAMTTNEE713 pKa = 4.4GQFHH717 pKa = 7.2LIDD720 pKa = 4.26AGGDD724 pKa = 3.63SGCTDD729 pKa = 2.95NDD731 pKa = 3.82ANVGGTEE738 pKa = 4.19VVSFMPEE745 pKa = 3.4EE746 pKa = 4.49LLPQLKK752 pKa = 9.35QVNGPITVEE761 pKa = 4.02GTGQTRR767 pKa = 11.84FNTYY771 pKa = 10.49GLDD774 pKa = 3.47GSPVVWRR781 pKa = 11.84YY782 pKa = 9.93DD783 pKa = 3.12ADD785 pKa = 4.68AGDD788 pKa = 3.62GTARR792 pKa = 11.84DD793 pKa = 3.71GVINYY798 pKa = 8.94GADD801 pKa = 3.33SDD803 pKa = 4.09EE804 pKa = 4.59DD805 pKa = 3.74FVYY808 pKa = 10.84LFLTQRR814 pKa = 11.84RR815 pKa = 11.84GGKK818 pKa = 9.85GIWALDD824 pKa = 3.34ITNPTSPEE832 pKa = 3.99LMWKK836 pKa = 9.39VWDD839 pKa = 4.14GDD841 pKa = 3.88LTSVNGTSAADD852 pKa = 3.57VYY854 pKa = 11.57SEE856 pKa = 5.77LGQTWSTPKK865 pKa = 9.25LTKK868 pKa = 10.35IKK870 pKa = 10.65DD871 pKa = 3.41GEE873 pKa = 4.08YY874 pKa = 10.77DD875 pKa = 3.09RR876 pKa = 11.84GVLVFGGGYY885 pKa = 10.73DD886 pKa = 3.71PDD888 pKa = 3.66QDD890 pKa = 3.8DD891 pKa = 5.14AYY893 pKa = 11.31SFSDD897 pKa = 3.36TQGRR901 pKa = 11.84AIYY904 pKa = 10.53VVDD907 pKa = 4.95AYY909 pKa = 9.81TGEE912 pKa = 4.28PLWTVSGEE920 pKa = 4.25AASTTPTQPSGGASGTMLIPEE941 pKa = 4.72MDD943 pKa = 3.33NSIPAEE949 pKa = 4.06VQPADD954 pKa = 3.9IDD956 pKa = 3.81SDD958 pKa = 4.41GYY960 pKa = 11.04LDD962 pKa = 4.94RR963 pKa = 11.84IYY965 pKa = 11.18AVDD968 pKa = 3.33VVGRR972 pKa = 11.84VFRR975 pKa = 11.84IDD977 pKa = 5.0FDD979 pKa = 3.67ILPNGDD985 pKa = 3.57STSTQMYY992 pKa = 8.52GGGMVAALNDD1002 pKa = 3.78SSCDD1006 pKa = 3.33EE1007 pKa = 4.24EE1008 pKa = 6.17APTDD1012 pKa = 4.01GVADD1016 pKa = 3.61ACHH1019 pKa = 6.02RR1020 pKa = 11.84RR1021 pKa = 11.84FYY1023 pKa = 10.94NKK1025 pKa = 9.32PDD1027 pKa = 3.26VAVMVGYY1034 pKa = 8.61PAAPYY1039 pKa = 9.18VQVAVGSGYY1048 pKa = 10.22RR1049 pKa = 11.84AWPASVSGIQDD1060 pKa = 3.31RR1061 pKa = 11.84FYY1063 pKa = 11.69VLFDD1067 pKa = 3.95EE1068 pKa = 5.37NVIDD1072 pKa = 4.33PLLSSVYY1079 pKa = 9.13GTGAGEE1085 pKa = 4.1QYY1087 pKa = 10.81HH1088 pKa = 5.12WTEE1091 pKa = 5.01DD1092 pKa = 3.24SDD1094 pKa = 4.56LANLTDD1100 pKa = 3.4IPGDD1104 pKa = 3.41VSDD1107 pKa = 4.3YY1108 pKa = 11.51SGAQTAAEE1116 pKa = 3.88TALGNKK1122 pKa = 8.96HH1123 pKa = 5.02GWYY1126 pKa = 10.35FNLEE1130 pKa = 4.14LTTDD1134 pKa = 3.15AGGTVTSHH1142 pKa = 6.36EE1143 pKa = 4.54KK1144 pKa = 10.62VLSDD1148 pKa = 3.17SVTFNGLILFTTYY1161 pKa = 9.86IRR1163 pKa = 11.84SSEE1166 pKa = 4.17DD1167 pKa = 3.2EE1168 pKa = 4.54ANVCEE1173 pKa = 4.29PSLGSGRR1180 pKa = 11.84FYY1182 pKa = 11.31AVNAFNGLPVGDD1194 pKa = 4.02INQGVFFTEE1203 pKa = 3.84DD1204 pKa = 2.94TARR1207 pKa = 11.84SDD1209 pKa = 3.27FSEE1212 pKa = 4.06ADD1214 pKa = 3.04RR1215 pKa = 11.84YY1216 pKa = 10.21MNLGRR1221 pKa = 11.84KK1222 pKa = 8.66GIPTTPQIILTDD1234 pKa = 3.56EE1235 pKa = 4.53GPRR1238 pKa = 11.84VIVTTEE1244 pKa = 4.69LIPEE1248 pKa = 4.43GLLDD1252 pKa = 4.61SDD1254 pKa = 5.07LFSKK1258 pKa = 10.21KK1259 pKa = 7.04WWLDD1263 pKa = 3.16AKK1265 pKa = 10.96
Molecular weight: 137.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1T2KXJ6|A0A1T2KXJ6_9GAMM Phage-tail_3 domain-containing protein OS=Solemya velesiana gill symbiont OX=1918948 GN=BOW51_02575 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 9.4VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84ARR41 pKa = 11.84LTPP44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 9.4VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84ARR41 pKa = 11.84LTPP44 pKa = 3.93
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
693604 |
31 |
15124 |
301.0 |
33.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.078 ± 0.056 | 0.995 ± 0.027 |
5.965 ± 0.113 | 7.245 ± 0.056 |
3.664 ± 0.036 | 7.841 ± 0.084 |
2.266 ± 0.034 | 5.786 ± 0.039 |
4.354 ± 0.07 | 10.475 ± 0.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.733 ± 0.046 | 3.323 ± 0.044 |
4.467 ± 0.044 | 3.858 ± 0.043 |
6.088 ± 0.097 | 5.866 ± 0.042 |
5.008 ± 0.092 | 7.148 ± 0.048 |
1.206 ± 0.021 | 2.634 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
