
Tenacibaculum todarodis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Tenacibaculum
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
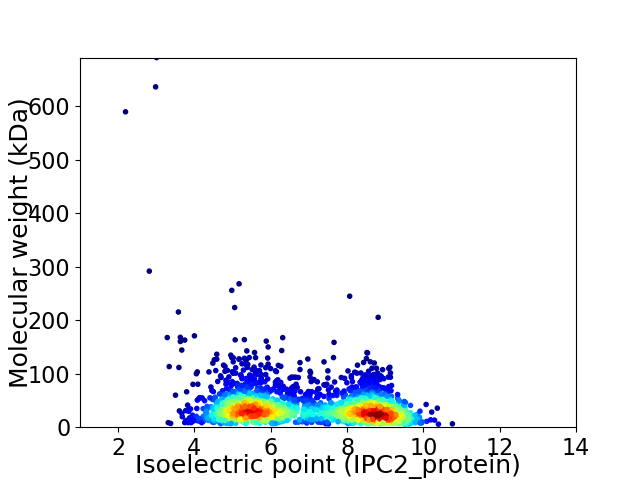
Virtual 2D-PAGE plot for 2636 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3JGQ9|A0A1L3JGQ9_9FLAO Uncharacterized protein OS=Tenacibaculum todarodis OX=1850252 GN=LPB136_02460 PE=4 SV=1
MM1 pKa = 7.59KK2 pKa = 10.37KK3 pKa = 10.18LLPLIFLFLTFNLFAQKK20 pKa = 9.71EE21 pKa = 4.13ANIWYY26 pKa = 9.23FGRR29 pKa = 11.84NAGIDD34 pKa = 3.78FSTTPPTALTDD45 pKa = 3.84GQLNTLEE52 pKa = 4.77GCSSFSDD59 pKa = 3.79SNGNLLFYY67 pKa = 11.05VGAPNSADD75 pKa = 3.13NRR77 pKa = 11.84LTVWDD82 pKa = 4.11KK83 pKa = 11.06NHH85 pKa = 7.17NIMNYY90 pKa = 8.77TNGNIGNNLLGHH102 pKa = 6.25TSSTQSAMIIPKK114 pKa = 9.87PEE116 pKa = 3.9STTIFYY122 pKa = 10.85LFTVDD127 pKa = 5.49DD128 pKa = 4.96GPGSGGAEE136 pKa = 3.81GMNLYY141 pKa = 9.5TIDD144 pKa = 3.42MSLNGGLGQLIDD156 pKa = 4.13EE157 pKa = 5.56DD158 pKa = 4.57GDD160 pKa = 3.85GDD162 pKa = 4.24FFEE165 pKa = 5.57DD166 pKa = 4.89LSDD169 pKa = 3.53IRR171 pKa = 11.84KK172 pKa = 9.56NDD174 pKa = 3.03WTEE177 pKa = 3.24KK178 pKa = 8.59VAAVRR183 pKa = 11.84GANCNEE189 pKa = 3.71FWVVSVVNNTFFSYY203 pKa = 10.38QVTSSGVNLTPVTSPVSTPAQRR225 pKa = 11.84RR226 pKa = 11.84GYY228 pKa = 10.71LKK230 pKa = 10.79LSPDD234 pKa = 3.37GSKK237 pKa = 10.66LAIANQDD244 pKa = 3.4SNAMVYY250 pKa = 10.63SFDD253 pKa = 3.56NSTGRR258 pKa = 11.84VSNDD262 pKa = 3.47GIILLNTGGDD272 pKa = 3.73GQPYY276 pKa = 9.56GVEE279 pKa = 4.03FSIDD283 pKa = 3.38SKK285 pKa = 11.32KK286 pKa = 11.01LYY288 pKa = 10.36FSTVSSFRR296 pKa = 11.84QDD298 pKa = 3.0LSEE301 pKa = 4.58PEE303 pKa = 3.52VTYY306 pKa = 11.04RR307 pKa = 11.84LFQFDD312 pKa = 5.94LIATDD317 pKa = 3.63IPGSKK322 pKa = 10.17SEE324 pKa = 4.33IYY326 pKa = 10.43RR327 pKa = 11.84NNNGFRR333 pKa = 11.84GALQLGPDD341 pKa = 3.48GRR343 pKa = 11.84IYY345 pKa = 10.56ATIPQAYY352 pKa = 9.11DD353 pKa = 3.65DD354 pKa = 4.79PNGDD358 pKa = 3.5ARR360 pKa = 11.84FLSVIEE366 pKa = 4.44NPTANATDD374 pKa = 4.14IIFTQNAIDD383 pKa = 3.78LGGRR387 pKa = 11.84TATQGLPPFISSLLLGIEE405 pKa = 4.43LTDD408 pKa = 3.37TTNNQIISDD417 pKa = 4.48EE418 pKa = 3.98IVQYY422 pKa = 11.12CLTEE426 pKa = 4.44NILIQPEE433 pKa = 4.22AVTPQTGTTVDD444 pKa = 4.01YY445 pKa = 10.7EE446 pKa = 4.21WTFDD450 pKa = 4.26DD451 pKa = 4.52GTTTSIISTTRR462 pKa = 11.84DD463 pKa = 3.02LTLNNIQFNNAGEE476 pKa = 4.19YY477 pKa = 9.64TLTVTLTDD485 pKa = 3.31EE486 pKa = 4.97CGNPTKK492 pKa = 11.03LEE494 pKa = 3.96GTFTIEE500 pKa = 4.17VYY502 pKa = 11.3NNLTPVAPTDD512 pKa = 4.44INFCDD517 pKa = 4.39DD518 pKa = 5.49DD519 pKa = 6.71GDD521 pKa = 4.03GLQAFDD527 pKa = 4.52FQTDD531 pKa = 3.96LTPTILNGQDD541 pKa = 3.2PTLVEE546 pKa = 3.92VVYY549 pKa = 11.1YY550 pKa = 8.24PTEE553 pKa = 3.86NDD555 pKa = 3.54ALNNTAPITNPFTPTAFYY573 pKa = 10.64DD574 pKa = 3.82GPIWVRR580 pKa = 11.84VHH582 pKa = 5.75NQRR585 pKa = 11.84APNACNEE592 pKa = 4.21VYY594 pKa = 10.72SFNLQITGEE603 pKa = 4.3PVAQTPDD610 pKa = 4.57DD611 pKa = 3.85IVACDD616 pKa = 3.66GDD618 pKa = 4.17GTDD621 pKa = 5.12SVDD624 pKa = 4.59NDD626 pKa = 3.37GLYY629 pKa = 11.1DD630 pKa = 3.82NFILNTRR637 pKa = 11.84DD638 pKa = 3.5AQILGALSEE647 pKa = 4.34TQYY650 pKa = 10.32TVSYY654 pKa = 8.11HH655 pKa = 5.96TSLADD660 pKa = 3.5AQTSATTNAIDD671 pKa = 4.85KK672 pKa = 7.54NTNYY676 pKa = 10.14QSGNDD681 pKa = 3.53IIFVRR686 pKa = 11.84VEE688 pKa = 3.63NVDD691 pKa = 3.59NVNCNDD697 pKa = 3.43TSQSFEE703 pKa = 5.16LIVNPLPTPVDD714 pKa = 3.74VTILQCDD721 pKa = 3.67DD722 pKa = 4.48DD723 pKa = 6.7ADD725 pKa = 4.06LVADD729 pKa = 3.96INLTLEE735 pKa = 4.1QEE737 pKa = 4.65NISTNHH743 pKa = 4.96TNEE746 pKa = 3.63TFEE749 pKa = 4.34YY750 pKa = 8.49FTTQAEE756 pKa = 4.75AIAGTPVIANPIAYY770 pKa = 8.22NASNGDD776 pKa = 3.78QVWVRR781 pKa = 11.84TISTEE786 pKa = 3.21NCYY789 pKa = 10.48RR790 pKa = 11.84ITTIDD795 pKa = 3.54IIISFASDD803 pKa = 3.07VPYY806 pKa = 10.68NRR808 pKa = 11.84NFIEE812 pKa = 5.29CDD814 pKa = 3.32DD815 pKa = 4.44FLDD818 pKa = 5.25LDD820 pKa = 4.54GNDD823 pKa = 3.61TPGANDD829 pKa = 3.75DD830 pKa = 3.82TDD832 pKa = 4.98GIATFDD838 pKa = 3.74FSAATSEE845 pKa = 4.34ILGHH849 pKa = 6.69PSLAGALNLDD859 pKa = 3.35VKK861 pKa = 10.89YY862 pKa = 10.83FEE864 pKa = 5.21TIADD868 pKa = 4.06RR869 pKa = 11.84ASEE872 pKa = 4.05INEE875 pKa = 3.53ISDD878 pKa = 3.86ISNHH882 pKa = 6.13RR883 pKa = 11.84NNNDD887 pKa = 3.37PAYY890 pKa = 10.63ANLQTIYY897 pKa = 11.0AKK899 pKa = 10.38IINTVNNDD907 pKa = 3.4CTGLAEE913 pKa = 4.68LTLQTTPVPVANAINPDD930 pKa = 3.61NEE932 pKa = 4.32YY933 pKa = 10.51EE934 pKa = 4.1LCDD937 pKa = 4.52DD938 pKa = 5.14FVDD941 pKa = 4.1ADD943 pKa = 3.87GSNGIIQSFDD953 pKa = 3.88LEE955 pKa = 4.45SQTAAILGSQDD966 pKa = 3.06PLAYY970 pKa = 9.46TITYY974 pKa = 9.58HH975 pKa = 7.06EE976 pKa = 4.53SAADD980 pKa = 3.61ATSGANPLASPYY992 pKa = 10.84TNITRR997 pKa = 11.84DD998 pKa = 3.09RR999 pKa = 11.84QTIYY1003 pKa = 11.26ARR1005 pKa = 11.84VSGNGCYY1012 pKa = 9.68FDD1014 pKa = 5.42RR1015 pKa = 11.84YY1016 pKa = 10.82SFDD1019 pKa = 2.99IVVNSLPIANQLNMANTLEE1038 pKa = 4.19VCDD1041 pKa = 5.14DD1042 pKa = 4.41DD1043 pKa = 7.03ADD1045 pKa = 4.1GSATNGFVQNIDD1057 pKa = 4.01LEE1059 pKa = 4.66TLTTTILGTQDD1070 pKa = 3.18PTNFTVTYY1078 pKa = 7.19HH1079 pKa = 5.98TSYY1082 pKa = 11.87ALAQTGAIPITGMYY1096 pKa = 10.63SNTTQGGDD1104 pKa = 3.14TVYY1107 pKa = 10.95VRR1109 pKa = 11.84IVNNTTTCVNDD1120 pKa = 3.49TFNFQVLINAEE1131 pKa = 4.09PLANPDD1137 pKa = 3.65NLVSNLSEE1145 pKa = 4.66CDD1147 pKa = 3.19NDD1149 pKa = 5.25ADD1151 pKa = 4.81GDD1153 pKa = 4.11DD1154 pKa = 4.32ANGFVQSFDD1163 pKa = 4.47LEE1165 pKa = 4.36SQIPDD1170 pKa = 3.08ILGDD1174 pKa = 3.53PTVQDD1179 pKa = 3.37EE1180 pKa = 4.74DD1181 pKa = 4.08DD1182 pKa = 4.39FNVTFHH1188 pKa = 6.79LTQADD1193 pKa = 3.92ATDD1196 pKa = 4.2GSAPQSSPFTNTDD1209 pKa = 2.6INQQTIFVRR1218 pKa = 11.84IEE1220 pKa = 3.74NKK1222 pKa = 9.25ATGCVNDD1229 pKa = 5.17DD1230 pKa = 3.54LTFDD1234 pKa = 4.17VIVNSLPDD1242 pKa = 3.68FVVTSPQIVCLNNPPLTLTVEE1263 pKa = 4.35NPNDD1267 pKa = 3.14VYY1269 pKa = 10.84TYY1271 pKa = 10.46QWFDD1275 pKa = 2.92ASGNPLGTAITQDD1288 pKa = 3.33VTVGGDD1294 pKa = 3.53YY1295 pKa = 11.01SVTATTTNGTMCPRR1309 pKa = 11.84TRR1311 pKa = 11.84TITVNEE1317 pKa = 4.23SNIATITDD1325 pKa = 3.7DD1326 pKa = 5.93DD1327 pKa = 3.81ITIVDD1332 pKa = 3.86DD1333 pKa = 4.41SEE1335 pKa = 4.67NNSITINDD1343 pKa = 3.89SNLGEE1348 pKa = 4.26GDD1350 pKa = 3.89YY1351 pKa = 11.29EE1352 pKa = 4.31FALIDD1357 pKa = 3.77EE1358 pKa = 4.75NGFTIVNYY1366 pKa = 9.71QDD1368 pKa = 3.31EE1369 pKa = 4.72AFFDD1373 pKa = 4.08GLNGGIYY1380 pKa = 9.41TIHH1383 pKa = 7.17IRR1385 pKa = 11.84DD1386 pKa = 3.61KK1387 pKa = 10.47NGCGVTTHH1395 pKa = 6.44TVPVIEE1401 pKa = 4.59FPKK1404 pKa = 10.59FFTPNNDD1411 pKa = 3.67GIKK1414 pKa = 10.46DD1415 pKa = 3.26VWTIKK1420 pKa = 9.08GTNATFFPINTVVIFNRR1437 pKa = 11.84QGKK1440 pKa = 9.7IINQFNINAYY1450 pKa = 9.53GWDD1453 pKa = 3.26GHH1455 pKa = 6.19YY1456 pKa = 10.1KK1457 pKa = 10.35DD1458 pKa = 5.42KK1459 pKa = 11.46AMPSNNYY1466 pKa = 7.47WFKK1469 pKa = 10.95ATLVDD1474 pKa = 3.57AKK1476 pKa = 10.84GITRR1480 pKa = 11.84TRR1482 pKa = 11.84SGYY1485 pKa = 9.96FSLIRR1490 pKa = 11.84RR1491 pKa = 4.0
MM1 pKa = 7.59KK2 pKa = 10.37KK3 pKa = 10.18LLPLIFLFLTFNLFAQKK20 pKa = 9.71EE21 pKa = 4.13ANIWYY26 pKa = 9.23FGRR29 pKa = 11.84NAGIDD34 pKa = 3.78FSTTPPTALTDD45 pKa = 3.84GQLNTLEE52 pKa = 4.77GCSSFSDD59 pKa = 3.79SNGNLLFYY67 pKa = 11.05VGAPNSADD75 pKa = 3.13NRR77 pKa = 11.84LTVWDD82 pKa = 4.11KK83 pKa = 11.06NHH85 pKa = 7.17NIMNYY90 pKa = 8.77TNGNIGNNLLGHH102 pKa = 6.25TSSTQSAMIIPKK114 pKa = 9.87PEE116 pKa = 3.9STTIFYY122 pKa = 10.85LFTVDD127 pKa = 5.49DD128 pKa = 4.96GPGSGGAEE136 pKa = 3.81GMNLYY141 pKa = 9.5TIDD144 pKa = 3.42MSLNGGLGQLIDD156 pKa = 4.13EE157 pKa = 5.56DD158 pKa = 4.57GDD160 pKa = 3.85GDD162 pKa = 4.24FFEE165 pKa = 5.57DD166 pKa = 4.89LSDD169 pKa = 3.53IRR171 pKa = 11.84KK172 pKa = 9.56NDD174 pKa = 3.03WTEE177 pKa = 3.24KK178 pKa = 8.59VAAVRR183 pKa = 11.84GANCNEE189 pKa = 3.71FWVVSVVNNTFFSYY203 pKa = 10.38QVTSSGVNLTPVTSPVSTPAQRR225 pKa = 11.84RR226 pKa = 11.84GYY228 pKa = 10.71LKK230 pKa = 10.79LSPDD234 pKa = 3.37GSKK237 pKa = 10.66LAIANQDD244 pKa = 3.4SNAMVYY250 pKa = 10.63SFDD253 pKa = 3.56NSTGRR258 pKa = 11.84VSNDD262 pKa = 3.47GIILLNTGGDD272 pKa = 3.73GQPYY276 pKa = 9.56GVEE279 pKa = 4.03FSIDD283 pKa = 3.38SKK285 pKa = 11.32KK286 pKa = 11.01LYY288 pKa = 10.36FSTVSSFRR296 pKa = 11.84QDD298 pKa = 3.0LSEE301 pKa = 4.58PEE303 pKa = 3.52VTYY306 pKa = 11.04RR307 pKa = 11.84LFQFDD312 pKa = 5.94LIATDD317 pKa = 3.63IPGSKK322 pKa = 10.17SEE324 pKa = 4.33IYY326 pKa = 10.43RR327 pKa = 11.84NNNGFRR333 pKa = 11.84GALQLGPDD341 pKa = 3.48GRR343 pKa = 11.84IYY345 pKa = 10.56ATIPQAYY352 pKa = 9.11DD353 pKa = 3.65DD354 pKa = 4.79PNGDD358 pKa = 3.5ARR360 pKa = 11.84FLSVIEE366 pKa = 4.44NPTANATDD374 pKa = 4.14IIFTQNAIDD383 pKa = 3.78LGGRR387 pKa = 11.84TATQGLPPFISSLLLGIEE405 pKa = 4.43LTDD408 pKa = 3.37TTNNQIISDD417 pKa = 4.48EE418 pKa = 3.98IVQYY422 pKa = 11.12CLTEE426 pKa = 4.44NILIQPEE433 pKa = 4.22AVTPQTGTTVDD444 pKa = 4.01YY445 pKa = 10.7EE446 pKa = 4.21WTFDD450 pKa = 4.26DD451 pKa = 4.52GTTTSIISTTRR462 pKa = 11.84DD463 pKa = 3.02LTLNNIQFNNAGEE476 pKa = 4.19YY477 pKa = 9.64TLTVTLTDD485 pKa = 3.31EE486 pKa = 4.97CGNPTKK492 pKa = 11.03LEE494 pKa = 3.96GTFTIEE500 pKa = 4.17VYY502 pKa = 11.3NNLTPVAPTDD512 pKa = 4.44INFCDD517 pKa = 4.39DD518 pKa = 5.49DD519 pKa = 6.71GDD521 pKa = 4.03GLQAFDD527 pKa = 4.52FQTDD531 pKa = 3.96LTPTILNGQDD541 pKa = 3.2PTLVEE546 pKa = 3.92VVYY549 pKa = 11.1YY550 pKa = 8.24PTEE553 pKa = 3.86NDD555 pKa = 3.54ALNNTAPITNPFTPTAFYY573 pKa = 10.64DD574 pKa = 3.82GPIWVRR580 pKa = 11.84VHH582 pKa = 5.75NQRR585 pKa = 11.84APNACNEE592 pKa = 4.21VYY594 pKa = 10.72SFNLQITGEE603 pKa = 4.3PVAQTPDD610 pKa = 4.57DD611 pKa = 3.85IVACDD616 pKa = 3.66GDD618 pKa = 4.17GTDD621 pKa = 5.12SVDD624 pKa = 4.59NDD626 pKa = 3.37GLYY629 pKa = 11.1DD630 pKa = 3.82NFILNTRR637 pKa = 11.84DD638 pKa = 3.5AQILGALSEE647 pKa = 4.34TQYY650 pKa = 10.32TVSYY654 pKa = 8.11HH655 pKa = 5.96TSLADD660 pKa = 3.5AQTSATTNAIDD671 pKa = 4.85KK672 pKa = 7.54NTNYY676 pKa = 10.14QSGNDD681 pKa = 3.53IIFVRR686 pKa = 11.84VEE688 pKa = 3.63NVDD691 pKa = 3.59NVNCNDD697 pKa = 3.43TSQSFEE703 pKa = 5.16LIVNPLPTPVDD714 pKa = 3.74VTILQCDD721 pKa = 3.67DD722 pKa = 4.48DD723 pKa = 6.7ADD725 pKa = 4.06LVADD729 pKa = 3.96INLTLEE735 pKa = 4.1QEE737 pKa = 4.65NISTNHH743 pKa = 4.96TNEE746 pKa = 3.63TFEE749 pKa = 4.34YY750 pKa = 8.49FTTQAEE756 pKa = 4.75AIAGTPVIANPIAYY770 pKa = 8.22NASNGDD776 pKa = 3.78QVWVRR781 pKa = 11.84TISTEE786 pKa = 3.21NCYY789 pKa = 10.48RR790 pKa = 11.84ITTIDD795 pKa = 3.54IIISFASDD803 pKa = 3.07VPYY806 pKa = 10.68NRR808 pKa = 11.84NFIEE812 pKa = 5.29CDD814 pKa = 3.32DD815 pKa = 4.44FLDD818 pKa = 5.25LDD820 pKa = 4.54GNDD823 pKa = 3.61TPGANDD829 pKa = 3.75DD830 pKa = 3.82TDD832 pKa = 4.98GIATFDD838 pKa = 3.74FSAATSEE845 pKa = 4.34ILGHH849 pKa = 6.69PSLAGALNLDD859 pKa = 3.35VKK861 pKa = 10.89YY862 pKa = 10.83FEE864 pKa = 5.21TIADD868 pKa = 4.06RR869 pKa = 11.84ASEE872 pKa = 4.05INEE875 pKa = 3.53ISDD878 pKa = 3.86ISNHH882 pKa = 6.13RR883 pKa = 11.84NNNDD887 pKa = 3.37PAYY890 pKa = 10.63ANLQTIYY897 pKa = 11.0AKK899 pKa = 10.38IINTVNNDD907 pKa = 3.4CTGLAEE913 pKa = 4.68LTLQTTPVPVANAINPDD930 pKa = 3.61NEE932 pKa = 4.32YY933 pKa = 10.51EE934 pKa = 4.1LCDD937 pKa = 4.52DD938 pKa = 5.14FVDD941 pKa = 4.1ADD943 pKa = 3.87GSNGIIQSFDD953 pKa = 3.88LEE955 pKa = 4.45SQTAAILGSQDD966 pKa = 3.06PLAYY970 pKa = 9.46TITYY974 pKa = 9.58HH975 pKa = 7.06EE976 pKa = 4.53SAADD980 pKa = 3.61ATSGANPLASPYY992 pKa = 10.84TNITRR997 pKa = 11.84DD998 pKa = 3.09RR999 pKa = 11.84QTIYY1003 pKa = 11.26ARR1005 pKa = 11.84VSGNGCYY1012 pKa = 9.68FDD1014 pKa = 5.42RR1015 pKa = 11.84YY1016 pKa = 10.82SFDD1019 pKa = 2.99IVVNSLPIANQLNMANTLEE1038 pKa = 4.19VCDD1041 pKa = 5.14DD1042 pKa = 4.41DD1043 pKa = 7.03ADD1045 pKa = 4.1GSATNGFVQNIDD1057 pKa = 4.01LEE1059 pKa = 4.66TLTTTILGTQDD1070 pKa = 3.18PTNFTVTYY1078 pKa = 7.19HH1079 pKa = 5.98TSYY1082 pKa = 11.87ALAQTGAIPITGMYY1096 pKa = 10.63SNTTQGGDD1104 pKa = 3.14TVYY1107 pKa = 10.95VRR1109 pKa = 11.84IVNNTTTCVNDD1120 pKa = 3.49TFNFQVLINAEE1131 pKa = 4.09PLANPDD1137 pKa = 3.65NLVSNLSEE1145 pKa = 4.66CDD1147 pKa = 3.19NDD1149 pKa = 5.25ADD1151 pKa = 4.81GDD1153 pKa = 4.11DD1154 pKa = 4.32ANGFVQSFDD1163 pKa = 4.47LEE1165 pKa = 4.36SQIPDD1170 pKa = 3.08ILGDD1174 pKa = 3.53PTVQDD1179 pKa = 3.37EE1180 pKa = 4.74DD1181 pKa = 4.08DD1182 pKa = 4.39FNVTFHH1188 pKa = 6.79LTQADD1193 pKa = 3.92ATDD1196 pKa = 4.2GSAPQSSPFTNTDD1209 pKa = 2.6INQQTIFVRR1218 pKa = 11.84IEE1220 pKa = 3.74NKK1222 pKa = 9.25ATGCVNDD1229 pKa = 5.17DD1230 pKa = 3.54LTFDD1234 pKa = 4.17VIVNSLPDD1242 pKa = 3.68FVVTSPQIVCLNNPPLTLTVEE1263 pKa = 4.35NPNDD1267 pKa = 3.14VYY1269 pKa = 10.84TYY1271 pKa = 10.46QWFDD1275 pKa = 2.92ASGNPLGTAITQDD1288 pKa = 3.33VTVGGDD1294 pKa = 3.53YY1295 pKa = 11.01SVTATTTNGTMCPRR1309 pKa = 11.84TRR1311 pKa = 11.84TITVNEE1317 pKa = 4.23SNIATITDD1325 pKa = 3.7DD1326 pKa = 5.93DD1327 pKa = 3.81ITIVDD1332 pKa = 3.86DD1333 pKa = 4.41SEE1335 pKa = 4.67NNSITINDD1343 pKa = 3.89SNLGEE1348 pKa = 4.26GDD1350 pKa = 3.89YY1351 pKa = 11.29EE1352 pKa = 4.31FALIDD1357 pKa = 3.77EE1358 pKa = 4.75NGFTIVNYY1366 pKa = 9.71QDD1368 pKa = 3.31EE1369 pKa = 4.72AFFDD1373 pKa = 4.08GLNGGIYY1380 pKa = 9.41TIHH1383 pKa = 7.17IRR1385 pKa = 11.84DD1386 pKa = 3.61KK1387 pKa = 10.47NGCGVTTHH1395 pKa = 6.44TVPVIEE1401 pKa = 4.59FPKK1404 pKa = 10.59FFTPNNDD1411 pKa = 3.67GIKK1414 pKa = 10.46DD1415 pKa = 3.26VWTIKK1420 pKa = 9.08GTNATFFPINTVVIFNRR1437 pKa = 11.84QGKK1440 pKa = 9.7IINQFNINAYY1450 pKa = 9.53GWDD1453 pKa = 3.26GHH1455 pKa = 6.19YY1456 pKa = 10.1KK1457 pKa = 10.35DD1458 pKa = 5.42KK1459 pKa = 11.46AMPSNNYY1466 pKa = 7.47WFKK1469 pKa = 10.95ATLVDD1474 pKa = 3.57AKK1476 pKa = 10.84GITRR1480 pKa = 11.84TRR1482 pKa = 11.84SGYY1485 pKa = 9.96FSLIRR1490 pKa = 11.84RR1491 pKa = 4.0
Molecular weight: 162.78 kDa
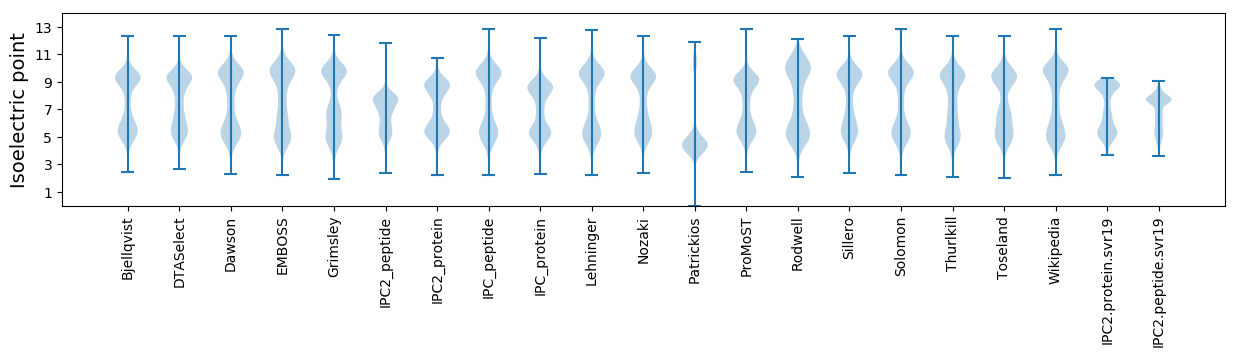
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3JFN5|A0A1L3JFN5_9FLAO Uncharacterized protein OS=Tenacibaculum todarodis OX=1850252 GN=LPB136_00550 PE=4 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.63LKK7 pKa = 10.48PVTPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.56KK28 pKa = 11.07PEE30 pKa = 4.39KK31 pKa = 10.58SLLAPNKK38 pKa = 10.02RR39 pKa = 11.84SGGRR43 pKa = 11.84NSQGRR48 pKa = 11.84MTTRR52 pKa = 11.84NIGGGHH58 pKa = 4.58KK59 pKa = 9.49QKK61 pKa = 10.8YY62 pKa = 9.64RR63 pKa = 11.84IIDD66 pKa = 3.67FKK68 pKa = 10.99KK69 pKa = 10.17DD70 pKa = 3.34KK71 pKa = 10.76QGIPATVKK79 pKa = 10.01TIEE82 pKa = 4.0YY83 pKa = 9.86DD84 pKa = 3.47PNRR87 pKa = 11.84TAFIALVFYY96 pKa = 11.16ADD98 pKa = 3.36GEE100 pKa = 4.23KK101 pKa = 10.28RR102 pKa = 11.84YY103 pKa = 10.74VIAQNGLKK111 pKa = 10.19VGQTIVSGRR120 pKa = 11.84EE121 pKa = 3.72ATPEE125 pKa = 3.74IGNALYY131 pKa = 10.83LSDD134 pKa = 4.56IPLGTTISCIEE145 pKa = 4.03LRR147 pKa = 11.84PGQGAVMARR156 pKa = 11.84SAGSFAQLMARR167 pKa = 11.84DD168 pKa = 3.96GKK170 pKa = 10.52YY171 pKa = 9.12ATVKK175 pKa = 10.42LPSGEE180 pKa = 4.06TRR182 pKa = 11.84LILLTCMATIGVVSNSDD199 pKa = 3.21HH200 pKa = 6.09QLLVSGKK207 pKa = 9.49AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TNPVRR224 pKa = 11.84MNPVDD229 pKa = 3.43HH230 pKa = 7.07PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.13PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.28GYY255 pKa = 7.21KK256 pKa = 8.8TRR258 pKa = 11.84SKK260 pKa = 9.63TKK262 pKa = 10.22ASNKK266 pKa = 9.95YY267 pKa = 9.56IIEE270 pKa = 4.06RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.1KK274 pKa = 9.82
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.63LKK7 pKa = 10.48PVTPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.56KK28 pKa = 11.07PEE30 pKa = 4.39KK31 pKa = 10.58SLLAPNKK38 pKa = 10.02RR39 pKa = 11.84SGGRR43 pKa = 11.84NSQGRR48 pKa = 11.84MTTRR52 pKa = 11.84NIGGGHH58 pKa = 4.58KK59 pKa = 9.49QKK61 pKa = 10.8YY62 pKa = 9.64RR63 pKa = 11.84IIDD66 pKa = 3.67FKK68 pKa = 10.99KK69 pKa = 10.17DD70 pKa = 3.34KK71 pKa = 10.76QGIPATVKK79 pKa = 10.01TIEE82 pKa = 4.0YY83 pKa = 9.86DD84 pKa = 3.47PNRR87 pKa = 11.84TAFIALVFYY96 pKa = 11.16ADD98 pKa = 3.36GEE100 pKa = 4.23KK101 pKa = 10.28RR102 pKa = 11.84YY103 pKa = 10.74VIAQNGLKK111 pKa = 10.19VGQTIVSGRR120 pKa = 11.84EE121 pKa = 3.72ATPEE125 pKa = 3.74IGNALYY131 pKa = 10.83LSDD134 pKa = 4.56IPLGTTISCIEE145 pKa = 4.03LRR147 pKa = 11.84PGQGAVMARR156 pKa = 11.84SAGSFAQLMARR167 pKa = 11.84DD168 pKa = 3.96GKK170 pKa = 10.52YY171 pKa = 9.12ATVKK175 pKa = 10.42LPSGEE180 pKa = 4.06TRR182 pKa = 11.84LILLTCMATIGVVSNSDD199 pKa = 3.21HH200 pKa = 6.09QLLVSGKK207 pKa = 9.49AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TNPVRR224 pKa = 11.84MNPVDD229 pKa = 3.43HH230 pKa = 7.07PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.13PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.28GYY255 pKa = 7.21KK256 pKa = 8.8TRR258 pKa = 11.84SKK260 pKa = 9.63TKK262 pKa = 10.22ASNKK266 pKa = 9.95YY267 pKa = 9.56IIEE270 pKa = 4.06RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.1KK274 pKa = 9.82
Molecular weight: 29.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
903262 |
50 |
6670 |
342.7 |
38.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.134 ± 0.055 | 0.686 ± 0.016 |
5.419 ± 0.079 | 6.669 ± 0.053 |
5.385 ± 0.069 | 6.204 ± 0.053 |
1.599 ± 0.029 | 8.137 ± 0.054 |
8.714 ± 0.112 | 9.063 ± 0.075 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.003 ± 0.034 | 6.674 ± 0.063 |
3.199 ± 0.039 | 3.221 ± 0.032 |
3.064 ± 0.044 | 6.506 ± 0.047 |
6.269 ± 0.166 | 6.285 ± 0.06 |
0.943 ± 0.019 | 3.824 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
