
Actinophytocola sp.
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Actinophytocola; unclassified Actinophytocola
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
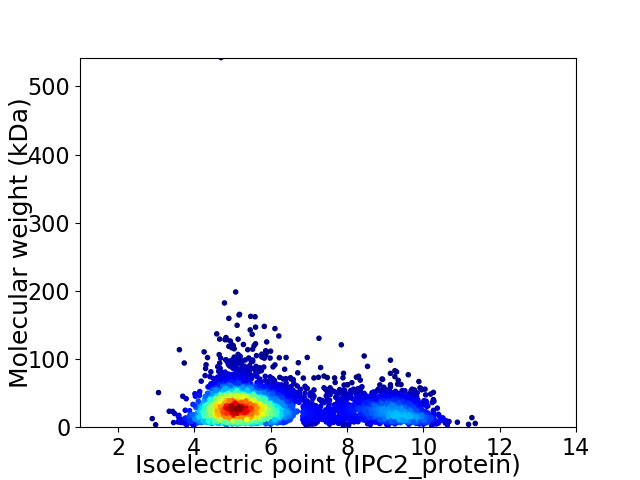
Virtual 2D-PAGE plot for 4585 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
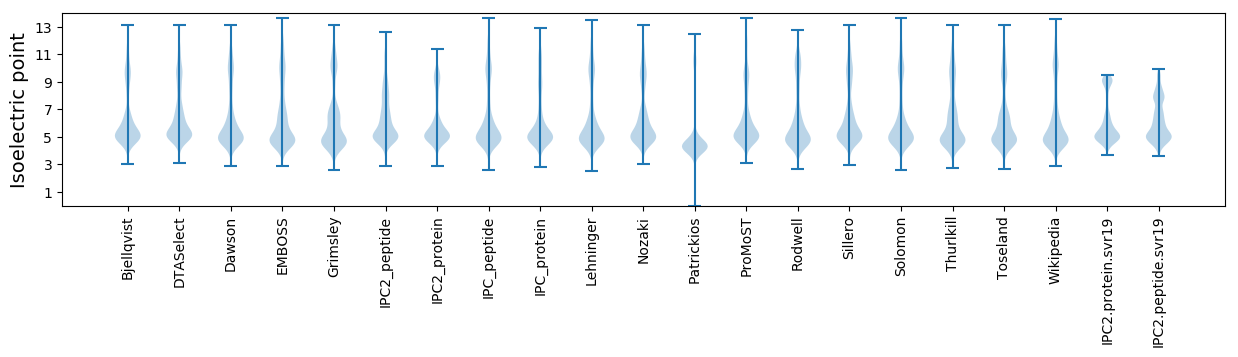
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6L5ESG1|A0A6L5ESG1_9PSEU Uncharacterized protein OS=Actinophytocola sp. OX=1872138 GN=GEV00_06790 PE=4 SV=1
MM1 pKa = 7.37RR2 pKa = 11.84RR3 pKa = 11.84KK4 pKa = 10.03FGVMLAACAMAGASVVSTQTATAEE28 pKa = 4.36TLGSEE33 pKa = 4.18LGSYY37 pKa = 9.88IVVLEE42 pKa = 4.79DD43 pKa = 3.56GVDD46 pKa = 3.45AASVAQQQVGALGGIVGHH64 pKa = 6.33VYY66 pKa = 10.72GSALSGYY73 pKa = 9.06SAQLTDD79 pKa = 3.42AARR82 pKa = 11.84DD83 pKa = 3.59RR84 pKa = 11.84LATYY88 pKa = 8.71PAVDD92 pKa = 4.57YY93 pKa = 10.62IQADD97 pKa = 3.98TPVRR101 pKa = 11.84ATQTMPTGIDD111 pKa = 3.06RR112 pKa = 11.84TDD114 pKa = 3.62AEE116 pKa = 4.4QSPTAAIDD124 pKa = 4.07GSDD127 pKa = 3.3EE128 pKa = 4.16RR129 pKa = 11.84VDD131 pKa = 3.29VDD133 pKa = 3.96VAVIDD138 pKa = 3.81TGADD142 pKa = 3.16MDD144 pKa = 4.61HH145 pKa = 7.6PDD147 pKa = 3.48LNVYY151 pKa = 10.05EE152 pKa = 4.91EE153 pKa = 4.43GAKK156 pKa = 10.25NCSTLSLSPEE166 pKa = 4.28DD167 pKa = 4.02EE168 pKa = 4.56NGHH171 pKa = 4.93GTHH174 pKa = 6.1VAGTIGALDD183 pKa = 4.13DD184 pKa = 4.05ASGVVGMAPGARR196 pKa = 11.84IWPVKK201 pKa = 10.08VLNALGMGLTSDD213 pKa = 4.38VICGIDD219 pKa = 3.47YY220 pKa = 10.77VAAHH224 pKa = 6.46SDD226 pKa = 3.83EE227 pKa = 5.01IEE229 pKa = 4.17VANMSLGGSGSDD241 pKa = 4.93DD242 pKa = 3.86GNCGEE247 pKa = 5.35DD248 pKa = 3.87NNDD251 pKa = 3.35AQHH254 pKa = 6.92AAICNAVDD262 pKa = 3.25AGVTFVVAAGNDD274 pKa = 3.51SADD277 pKa = 3.45ARR279 pKa = 11.84EE280 pKa = 4.4TVPAAYY286 pKa = 10.17DD287 pKa = 3.48EE288 pKa = 4.97VITVSALADD297 pKa = 3.69FDD299 pKa = 4.08GQPGGRR305 pKa = 11.84AAPTCRR311 pKa = 11.84DD312 pKa = 3.7DD313 pKa = 5.24VDD315 pKa = 3.87DD316 pKa = 4.32TFADD320 pKa = 4.97FSNYY324 pKa = 10.09GDD326 pKa = 5.33DD327 pKa = 4.04IDD329 pKa = 5.06VIAPGVCIEE338 pKa = 4.16STSLGGGYY346 pKa = 7.61EE347 pKa = 4.28TLSGTSMASPHH358 pKa = 6.26AAGAAALYY366 pKa = 9.07QANNPAASPEE376 pKa = 4.12QVKK379 pKa = 10.21RR380 pKa = 11.84ALQNAGTTDD389 pKa = 2.91WTFPSEE395 pKa = 4.2DD396 pKa = 3.36RR397 pKa = 11.84DD398 pKa = 4.4GIQEE402 pKa = 3.98MLVNVAGFF410 pKa = 3.38
MM1 pKa = 7.37RR2 pKa = 11.84RR3 pKa = 11.84KK4 pKa = 10.03FGVMLAACAMAGASVVSTQTATAEE28 pKa = 4.36TLGSEE33 pKa = 4.18LGSYY37 pKa = 9.88IVVLEE42 pKa = 4.79DD43 pKa = 3.56GVDD46 pKa = 3.45AASVAQQQVGALGGIVGHH64 pKa = 6.33VYY66 pKa = 10.72GSALSGYY73 pKa = 9.06SAQLTDD79 pKa = 3.42AARR82 pKa = 11.84DD83 pKa = 3.59RR84 pKa = 11.84LATYY88 pKa = 8.71PAVDD92 pKa = 4.57YY93 pKa = 10.62IQADD97 pKa = 3.98TPVRR101 pKa = 11.84ATQTMPTGIDD111 pKa = 3.06RR112 pKa = 11.84TDD114 pKa = 3.62AEE116 pKa = 4.4QSPTAAIDD124 pKa = 4.07GSDD127 pKa = 3.3EE128 pKa = 4.16RR129 pKa = 11.84VDD131 pKa = 3.29VDD133 pKa = 3.96VAVIDD138 pKa = 3.81TGADD142 pKa = 3.16MDD144 pKa = 4.61HH145 pKa = 7.6PDD147 pKa = 3.48LNVYY151 pKa = 10.05EE152 pKa = 4.91EE153 pKa = 4.43GAKK156 pKa = 10.25NCSTLSLSPEE166 pKa = 4.28DD167 pKa = 4.02EE168 pKa = 4.56NGHH171 pKa = 4.93GTHH174 pKa = 6.1VAGTIGALDD183 pKa = 4.13DD184 pKa = 4.05ASGVVGMAPGARR196 pKa = 11.84IWPVKK201 pKa = 10.08VLNALGMGLTSDD213 pKa = 4.38VICGIDD219 pKa = 3.47YY220 pKa = 10.77VAAHH224 pKa = 6.46SDD226 pKa = 3.83EE227 pKa = 5.01IEE229 pKa = 4.17VANMSLGGSGSDD241 pKa = 4.93DD242 pKa = 3.86GNCGEE247 pKa = 5.35DD248 pKa = 3.87NNDD251 pKa = 3.35AQHH254 pKa = 6.92AAICNAVDD262 pKa = 3.25AGVTFVVAAGNDD274 pKa = 3.51SADD277 pKa = 3.45ARR279 pKa = 11.84EE280 pKa = 4.4TVPAAYY286 pKa = 10.17DD287 pKa = 3.48EE288 pKa = 4.97VITVSALADD297 pKa = 3.69FDD299 pKa = 4.08GQPGGRR305 pKa = 11.84AAPTCRR311 pKa = 11.84DD312 pKa = 3.7DD313 pKa = 5.24VDD315 pKa = 3.87DD316 pKa = 4.32TFADD320 pKa = 4.97FSNYY324 pKa = 10.09GDD326 pKa = 5.33DD327 pKa = 4.04IDD329 pKa = 5.06VIAPGVCIEE338 pKa = 4.16STSLGGGYY346 pKa = 7.61EE347 pKa = 4.28TLSGTSMASPHH358 pKa = 6.26AAGAAALYY366 pKa = 9.07QANNPAASPEE376 pKa = 4.12QVKK379 pKa = 10.21RR380 pKa = 11.84ALQNAGTTDD389 pKa = 2.91WTFPSEE395 pKa = 4.2DD396 pKa = 3.36RR397 pKa = 11.84DD398 pKa = 4.4GIQEE402 pKa = 3.98MLVNVAGFF410 pKa = 3.38
Molecular weight: 41.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6L5ERD4|A0A6L5ERD4_9PSEU YCII domain-containing protein OS=Actinophytocola sp. OX=1872138 GN=GEV00_04855 PE=3 SV=1
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84SRR17 pKa = 11.84KK18 pKa = 8.57HH19 pKa = 4.77GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AILAARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.74GRR42 pKa = 11.84AKK44 pKa = 10.68LSAA47 pKa = 3.92
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84SRR17 pKa = 11.84KK18 pKa = 8.57HH19 pKa = 4.77GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AILAARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.74GRR42 pKa = 11.84AKK44 pKa = 10.68LSAA47 pKa = 3.92
Molecular weight: 5.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1350462 |
17 |
5027 |
294.5 |
31.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.078 ± 0.045 | 0.829 ± 0.01 |
6.396 ± 0.032 | 5.844 ± 0.036 |
2.77 ± 0.021 | 9.081 ± 0.033 |
2.316 ± 0.018 | 3.749 ± 0.025 |
2.105 ± 0.023 | 10.177 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.862 ± 0.013 | 1.884 ± 0.019 |
5.648 ± 0.027 | 2.818 ± 0.023 |
7.98 ± 0.04 | 5.119 ± 0.021 |
6.052 ± 0.025 | 8.8 ± 0.034 |
1.441 ± 0.016 | 2.049 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
