
Chlorella sorokiniana (Freshwater green alga)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; core chlorophytes; Trebouxiophyceae; Chlorellales; Chlorellaceae; Chlorella clade; Chlorella
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
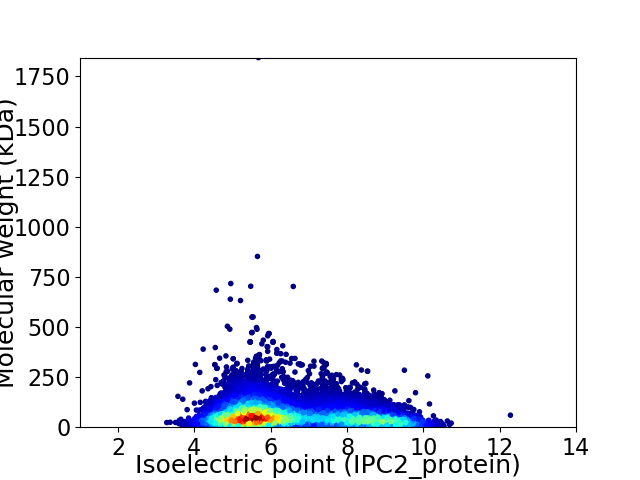
Virtual 2D-PAGE plot for 10201 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
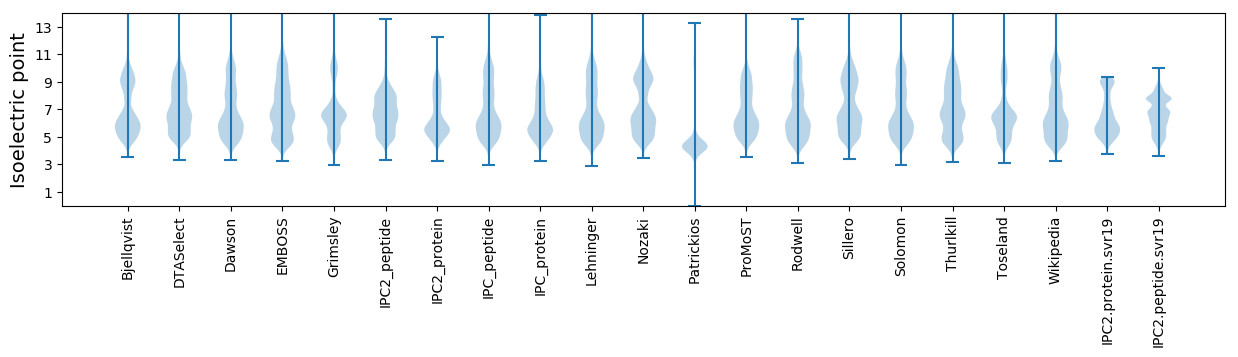
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P6U2K8|A0A2P6U2K8_CHLSO Ankyrin repeat isoform C OS=Chlorella sorokiniana OX=3076 GN=C2E21_1100 PE=3 SV=1
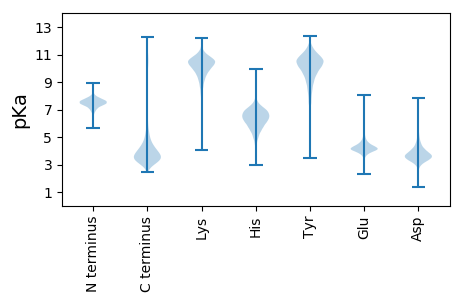
MM1 pKa = 6.92VAALVAGGADD11 pKa = 2.75INAPVEE17 pKa = 4.29ADD19 pKa = 3.57PCHH22 pKa = 6.68LPAYY26 pKa = 8.71TLLHH30 pKa = 5.65WAAYY34 pKa = 9.66SNAVDD39 pKa = 5.28AIQVLAACGADD50 pKa = 3.59LEE52 pKa = 4.79VRR54 pKa = 11.84DD55 pKa = 4.1ATVDD59 pKa = 3.45NSSLGRR65 pKa = 11.84WTEE68 pKa = 3.9FVGGGQTPLMVAAANGHH85 pKa = 5.13VEE87 pKa = 4.02AVEE90 pKa = 3.8ALIAAGCNANAGTTDD105 pKa = 3.06WGSWSPKK112 pKa = 9.54GATALEE118 pKa = 4.32FAVVGPGSGCQPGAQNPEE136 pKa = 3.88GLPAVVNALLAAGAVDD152 pKa = 3.72VEE154 pKa = 4.63YY155 pKa = 10.99ALEE158 pKa = 4.36AALHH162 pKa = 5.3RR163 pKa = 11.84HH164 pKa = 5.67ASEE167 pKa = 4.03EE168 pKa = 3.81MLAALEE174 pKa = 4.25AAVDD178 pKa = 3.9DD179 pKa = 5.23DD180 pKa = 4.78SEE182 pKa = 4.33EE183 pKa = 5.15DD184 pKa = 3.44EE185 pKa = 5.37DD186 pKa = 4.57EE187 pKa = 4.48EE188 pKa = 5.4SGSDD192 pKa = 3.51SDD194 pKa = 4.24
MM1 pKa = 6.92VAALVAGGADD11 pKa = 2.75INAPVEE17 pKa = 4.29ADD19 pKa = 3.57PCHH22 pKa = 6.68LPAYY26 pKa = 8.71TLLHH30 pKa = 5.65WAAYY34 pKa = 9.66SNAVDD39 pKa = 5.28AIQVLAACGADD50 pKa = 3.59LEE52 pKa = 4.79VRR54 pKa = 11.84DD55 pKa = 4.1ATVDD59 pKa = 3.45NSSLGRR65 pKa = 11.84WTEE68 pKa = 3.9FVGGGQTPLMVAAANGHH85 pKa = 5.13VEE87 pKa = 4.02AVEE90 pKa = 3.8ALIAAGCNANAGTTDD105 pKa = 3.06WGSWSPKK112 pKa = 9.54GATALEE118 pKa = 4.32FAVVGPGSGCQPGAQNPEE136 pKa = 3.88GLPAVVNALLAAGAVDD152 pKa = 3.72VEE154 pKa = 4.63YY155 pKa = 10.99ALEE158 pKa = 4.36AALHH162 pKa = 5.3RR163 pKa = 11.84HH164 pKa = 5.67ASEE167 pKa = 4.03EE168 pKa = 3.81MLAALEE174 pKa = 4.25AAVDD178 pKa = 3.9DD179 pKa = 5.23DD180 pKa = 4.78SEE182 pKa = 4.33EE183 pKa = 5.15DD184 pKa = 3.44EE185 pKa = 5.37DD186 pKa = 4.57EE187 pKa = 4.48EE188 pKa = 5.4SGSDD192 pKa = 3.51SDD194 pKa = 4.24
Molecular weight: 19.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P6TEX9|A0A2P6TEX9_CHLSO Metallothiol transferase isoform B OS=Chlorella sorokiniana OX=3076 GN=C2E21_8300 PE=4 SV=1
MM1 pKa = 7.48AALAPVQSLLHH12 pKa = 6.63LDD14 pKa = 3.63NALVAIPEE22 pKa = 4.27DD23 pKa = 3.88AACCSTSQPAAPARR37 pKa = 11.84WVDD40 pKa = 3.5HH41 pKa = 6.12EE42 pKa = 4.46RR43 pKa = 11.84LLAAGEE49 pKa = 3.97ARR51 pKa = 11.84VRR53 pKa = 11.84PLRR56 pKa = 11.84KK57 pKa = 9.17CASVPTLHH65 pKa = 7.03SMQEE69 pKa = 3.81ACGEE73 pKa = 4.29SEE75 pKa = 3.84GMEE78 pKa = 4.15VYY80 pKa = 10.76VILRR84 pKa = 11.84PFKK87 pKa = 10.56EE88 pKa = 4.0FGGGLFRR95 pKa = 11.84RR96 pKa = 11.84LPKK99 pKa = 9.89RR100 pKa = 11.84VKK102 pKa = 10.71NGVRR106 pKa = 11.84DD107 pKa = 3.74CGLCHH112 pKa = 6.21YY113 pKa = 10.44LAVFKK118 pKa = 11.05QKK120 pKa = 10.66DD121 pKa = 3.48GSLVQFDD128 pKa = 4.0FGPRR132 pKa = 11.84GGDD135 pKa = 2.76IHH137 pKa = 6.42VARR140 pKa = 11.84GPFAFLSKK148 pKa = 10.71SADD151 pKa = 2.87GKK153 pKa = 7.61MQRR156 pKa = 11.84LVPGEE161 pKa = 3.84VRR163 pKa = 11.84EE164 pKa = 4.01RR165 pKa = 11.84RR166 pKa = 11.84LMRR169 pKa = 11.84LPDD172 pKa = 2.78AHH174 pKa = 6.57MYY176 pKa = 9.76VGRR179 pKa = 11.84TPLSLEE185 pKa = 5.81DD186 pKa = 3.2IRR188 pKa = 11.84AWNALQEE195 pKa = 4.31QGSMHH200 pKa = 6.49YY201 pKa = 9.63EE202 pKa = 3.8LHH204 pKa = 7.27RR205 pKa = 11.84NDD207 pKa = 3.07CRR209 pKa = 11.84HH210 pKa = 6.43YY211 pKa = 10.22INCLVKK217 pKa = 9.05YY218 pKa = 5.39TTGRR222 pKa = 11.84EE223 pKa = 3.96QAASACLAAQWQRR236 pKa = 11.84ARR238 pKa = 11.84QLGTYY243 pKa = 9.94GLATSVVRR251 pKa = 11.84LTQFFTDD258 pKa = 4.1LANWGKK264 pKa = 9.61VQLIGNVSMYY274 pKa = 11.33GMMALSGQKK283 pKa = 9.92VLARR287 pKa = 11.84LPALLPGAKK296 pKa = 9.89AKK298 pKa = 9.62LQPVVSKK305 pKa = 11.05ALAGGVKK312 pKa = 9.92RR313 pKa = 11.84ALTGPVRR320 pKa = 11.84TAIARR325 pKa = 11.84KK326 pKa = 7.93PVVVGTAAMATLAASSSQAPMIKK349 pKa = 8.81EE350 pKa = 4.31TVSVGARR357 pKa = 11.84VARR360 pKa = 11.84AAQATIAAASTAVTRR375 pKa = 11.84ASAATVRR382 pKa = 11.84TTSQAAVAVGSITRR396 pKa = 11.84GAVALAAGRR405 pKa = 11.84PMPLALEE412 pKa = 4.43WGRR415 pKa = 11.84AASPPRR421 pKa = 11.84ASLGMRR427 pKa = 11.84AADD430 pKa = 3.62RR431 pKa = 11.84SQRR434 pKa = 11.84LALAITSAARR444 pKa = 11.84RR445 pKa = 3.73
MM1 pKa = 7.48AALAPVQSLLHH12 pKa = 6.63LDD14 pKa = 3.63NALVAIPEE22 pKa = 4.27DD23 pKa = 3.88AACCSTSQPAAPARR37 pKa = 11.84WVDD40 pKa = 3.5HH41 pKa = 6.12EE42 pKa = 4.46RR43 pKa = 11.84LLAAGEE49 pKa = 3.97ARR51 pKa = 11.84VRR53 pKa = 11.84PLRR56 pKa = 11.84KK57 pKa = 9.17CASVPTLHH65 pKa = 7.03SMQEE69 pKa = 3.81ACGEE73 pKa = 4.29SEE75 pKa = 3.84GMEE78 pKa = 4.15VYY80 pKa = 10.76VILRR84 pKa = 11.84PFKK87 pKa = 10.56EE88 pKa = 4.0FGGGLFRR95 pKa = 11.84RR96 pKa = 11.84LPKK99 pKa = 9.89RR100 pKa = 11.84VKK102 pKa = 10.71NGVRR106 pKa = 11.84DD107 pKa = 3.74CGLCHH112 pKa = 6.21YY113 pKa = 10.44LAVFKK118 pKa = 11.05QKK120 pKa = 10.66DD121 pKa = 3.48GSLVQFDD128 pKa = 4.0FGPRR132 pKa = 11.84GGDD135 pKa = 2.76IHH137 pKa = 6.42VARR140 pKa = 11.84GPFAFLSKK148 pKa = 10.71SADD151 pKa = 2.87GKK153 pKa = 7.61MQRR156 pKa = 11.84LVPGEE161 pKa = 3.84VRR163 pKa = 11.84EE164 pKa = 4.01RR165 pKa = 11.84RR166 pKa = 11.84LMRR169 pKa = 11.84LPDD172 pKa = 2.78AHH174 pKa = 6.57MYY176 pKa = 9.76VGRR179 pKa = 11.84TPLSLEE185 pKa = 5.81DD186 pKa = 3.2IRR188 pKa = 11.84AWNALQEE195 pKa = 4.31QGSMHH200 pKa = 6.49YY201 pKa = 9.63EE202 pKa = 3.8LHH204 pKa = 7.27RR205 pKa = 11.84NDD207 pKa = 3.07CRR209 pKa = 11.84HH210 pKa = 6.43YY211 pKa = 10.22INCLVKK217 pKa = 9.05YY218 pKa = 5.39TTGRR222 pKa = 11.84EE223 pKa = 3.96QAASACLAAQWQRR236 pKa = 11.84ARR238 pKa = 11.84QLGTYY243 pKa = 9.94GLATSVVRR251 pKa = 11.84LTQFFTDD258 pKa = 4.1LANWGKK264 pKa = 9.61VQLIGNVSMYY274 pKa = 11.33GMMALSGQKK283 pKa = 9.92VLARR287 pKa = 11.84LPALLPGAKK296 pKa = 9.89AKK298 pKa = 9.62LQPVVSKK305 pKa = 11.05ALAGGVKK312 pKa = 9.92RR313 pKa = 11.84ALTGPVRR320 pKa = 11.84TAIARR325 pKa = 11.84KK326 pKa = 7.93PVVVGTAAMATLAASSSQAPMIKK349 pKa = 8.81EE350 pKa = 4.31TVSVGARR357 pKa = 11.84VARR360 pKa = 11.84AAQATIAAASTAVTRR375 pKa = 11.84ASAATVRR382 pKa = 11.84TTSQAAVAVGSITRR396 pKa = 11.84GAVALAAGRR405 pKa = 11.84PMPLALEE412 pKa = 4.43WGRR415 pKa = 11.84AASPPRR421 pKa = 11.84ASLGMRR427 pKa = 11.84AADD430 pKa = 3.62RR431 pKa = 11.84SQRR434 pKa = 11.84LALAITSAARR444 pKa = 11.84RR445 pKa = 3.73
Molecular weight: 47.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
6715017 |
54 |
18393 |
658.3 |
69.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.604 ± 0.05 | 1.646 ± 0.015 |
4.438 ± 0.013 | 5.941 ± 0.026 |
2.63 ± 0.012 | 8.821 ± 0.022 |
2.036 ± 0.01 | 2.523 ± 0.014 |
3.157 ± 0.019 | 10.146 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.895 ± 0.008 | 2.043 ± 0.012 |
6.398 ± 0.027 | 5.682 ± 0.028 |
6.359 ± 0.02 | 6.759 ± 0.02 |
4.428 ± 0.026 | 6.162 ± 0.017 |
1.416 ± 0.008 | 1.917 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
