
Rhodobacter veldkampii DSM 11550
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Bieblia; Bieblia veldkampii
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
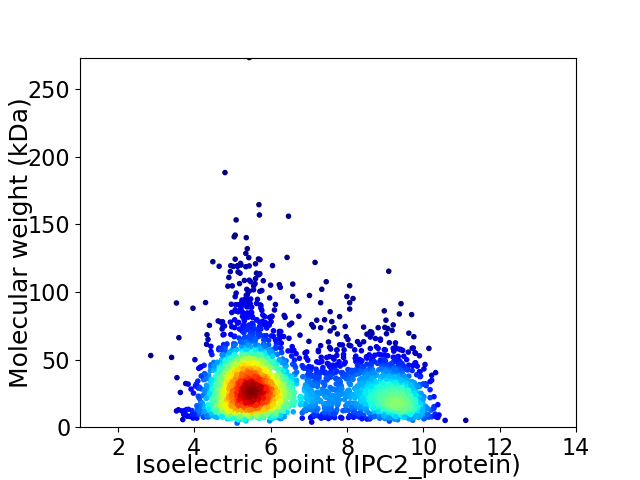
Virtual 2D-PAGE plot for 3115 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T4JL83|A0A2T4JL83_9RHOB Alpha-ketoglutarate dehydrogenase OS=Rhodobacter veldkampii DSM 11550 OX=1185920 GN=C5F46_03440 PE=4 SV=1
MM1 pKa = 8.11RR2 pKa = 11.84EE3 pKa = 3.93GDD5 pKa = 4.12DD6 pKa = 4.01LQIMLADD13 pKa = 3.72GRR15 pKa = 11.84TITLSGYY22 pKa = 9.45YY23 pKa = 9.44DD24 pKa = 3.27AHH26 pKa = 5.93ARR28 pKa = 11.84LYY30 pKa = 10.29ISSDD34 pKa = 3.57GEE36 pKa = 4.16LTPVTLEE43 pKa = 4.23DD44 pKa = 3.75GGNGVIYY51 pKa = 10.47ADD53 pKa = 3.62YY54 pKa = 10.53GQVEE58 pKa = 5.18VIGKK62 pKa = 8.39WSPNDD67 pKa = 3.12QLAFLGGDD75 pKa = 4.11EE76 pKa = 4.4ILSPAGDD83 pKa = 4.26DD84 pKa = 3.72TTGMAMFAPAMLGMGGLGAAGAGLAGLGLLGLGGGGSKK122 pKa = 10.79GGGDD126 pKa = 3.76DD127 pKa = 3.56TTPGGGEE134 pKa = 3.9VDD136 pKa = 4.54PIVPTVDD143 pKa = 3.2TPDD146 pKa = 3.37AQTTLTTNTEE156 pKa = 3.95DD157 pKa = 3.41PAATVSGTGEE167 pKa = 4.0PGSTVEE173 pKa = 5.4VVLGDD178 pKa = 3.48QTLEE182 pKa = 4.02TTIDD186 pKa = 3.67EE187 pKa = 4.87DD188 pKa = 4.39GTWSVTFEE196 pKa = 5.27DD197 pKa = 4.46DD198 pKa = 3.67TFPSDD203 pKa = 3.8GDD205 pKa = 3.68LTSTVDD211 pKa = 3.12VTAPDD216 pKa = 3.61GTEE219 pKa = 3.91YY220 pKa = 10.86TLDD223 pKa = 4.02GPDD226 pKa = 4.19FLIDD230 pKa = 3.52MTPPAVVITEE240 pKa = 4.54GALSTGDD247 pKa = 3.51VEE249 pKa = 5.62NLSEE253 pKa = 4.25YY254 pKa = 10.56QDD256 pKa = 4.35GITVRR261 pKa = 11.84GTGEE265 pKa = 3.61AGAAIVVAVAGHH277 pKa = 6.07AQATTVAADD286 pKa = 4.21GSWSVTFTTEE296 pKa = 3.57QMPAGTYY303 pKa = 9.87SEE305 pKa = 4.68AMTVTATDD313 pKa = 3.11AVGNVTTLNDD323 pKa = 3.66TLVVDD328 pKa = 4.8TEE330 pKa = 4.25ASVAFSSASVTGDD343 pKa = 3.36NIVNADD349 pKa = 3.56EE350 pKa = 4.54AQTGFALSGTSEE362 pKa = 4.25PGSVSVRR369 pKa = 11.84IEE371 pKa = 3.94LGGTSYY377 pKa = 11.36SATPAADD384 pKa = 3.93GTWSIALQDD393 pKa = 3.54VALATGVHH401 pKa = 5.79TATVTATDD409 pKa = 3.81ANGNTASATRR419 pKa = 11.84AISFDD424 pKa = 3.32TDD426 pKa = 3.29TSVSFDD432 pKa = 3.19SAQAGGNNLLNAAEE446 pKa = 4.14RR447 pKa = 11.84SAGLVLTGTAEE458 pKa = 4.07PGATVVVAFEE468 pKa = 4.34QGSHH472 pKa = 4.92TVTAGADD479 pKa = 3.91GHH481 pKa = 6.36WSASFAASEE490 pKa = 4.34VRR492 pKa = 11.84TGTYY496 pKa = 10.45SSTATVTATDD506 pKa = 3.11IAGNVATASHH516 pKa = 7.51TINVDD521 pKa = 3.4TEE523 pKa = 4.52VQNFTHH529 pKa = 6.99TSPTANAIVADD540 pKa = 4.75DD541 pKa = 3.47IVNARR546 pKa = 11.84EE547 pKa = 3.83AAGGLVVTGTVEE559 pKa = 3.84PGATVTVQLASGSVIPAINLGNGSWTATIPAAQLPAVEE597 pKa = 4.54TDD599 pKa = 3.1NVTLTVRR606 pKa = 11.84ATDD609 pKa = 3.59VNGNTATQTSSIDD622 pKa = 3.34FDD624 pKa = 3.78PVVRR628 pKa = 11.84NFAPNATVAGDD639 pKa = 4.26GIVNAQEE646 pKa = 4.19GADD649 pKa = 3.53GFVISGAVEE658 pKa = 3.87PGSRR662 pKa = 11.84VAVTLASGAVEE673 pKa = 4.2TVTAGANGQWSVMFHH688 pKa = 6.72SDD690 pKa = 4.08DD691 pKa = 4.7LGTTGSGTMGYY702 pKa = 9.85QIAATDD708 pKa = 3.8PVGNSATASGSFGYY722 pKa = 10.7DD723 pKa = 2.91LVAPEE728 pKa = 4.93SPDD731 pKa = 3.12IIAFTRR737 pKa = 11.84DD738 pKa = 3.03AGSLLGIRR746 pKa = 11.84TDD748 pKa = 3.53LGEE751 pKa = 4.04NTYY754 pKa = 10.76EE755 pKa = 3.96IAAVDD760 pKa = 3.19AAGRR764 pKa = 11.84VTGVPNDD771 pKa = 3.84ASVSTHH777 pKa = 5.74GGYY780 pKa = 8.1GTYY783 pKa = 10.62DD784 pKa = 4.24FYY786 pKa = 11.99SSVPNGSYY794 pKa = 10.65LVVSDD799 pKa = 4.79HH800 pKa = 6.74DD801 pKa = 3.89TAGNEE806 pKa = 4.0TSTLLVVDD814 pKa = 3.68NTSAVTVDD822 pKa = 4.12LARR825 pKa = 11.84SGLGDD830 pKa = 3.83FDD832 pKa = 5.64FGTIDD837 pKa = 3.47LTFAPEE843 pKa = 3.78AQLTITEE850 pKa = 4.21AQLVALTGPDD860 pKa = 3.35HH861 pKa = 7.0ALVIQGDD868 pKa = 4.28SADD871 pKa = 3.76HH872 pKa = 5.09VTALGAEE879 pKa = 4.22DD880 pKa = 4.16SGQNTVIGGHH890 pKa = 5.67TYY892 pKa = 10.59SVYY895 pKa = 10.76TLGDD899 pKa = 3.87DD900 pKa = 3.97GASLIIDD907 pKa = 3.84DD908 pKa = 5.07QITHH912 pKa = 6.02LTII915 pKa = 4.16
MM1 pKa = 8.11RR2 pKa = 11.84EE3 pKa = 3.93GDD5 pKa = 4.12DD6 pKa = 4.01LQIMLADD13 pKa = 3.72GRR15 pKa = 11.84TITLSGYY22 pKa = 9.45YY23 pKa = 9.44DD24 pKa = 3.27AHH26 pKa = 5.93ARR28 pKa = 11.84LYY30 pKa = 10.29ISSDD34 pKa = 3.57GEE36 pKa = 4.16LTPVTLEE43 pKa = 4.23DD44 pKa = 3.75GGNGVIYY51 pKa = 10.47ADD53 pKa = 3.62YY54 pKa = 10.53GQVEE58 pKa = 5.18VIGKK62 pKa = 8.39WSPNDD67 pKa = 3.12QLAFLGGDD75 pKa = 4.11EE76 pKa = 4.4ILSPAGDD83 pKa = 4.26DD84 pKa = 3.72TTGMAMFAPAMLGMGGLGAAGAGLAGLGLLGLGGGGSKK122 pKa = 10.79GGGDD126 pKa = 3.76DD127 pKa = 3.56TTPGGGEE134 pKa = 3.9VDD136 pKa = 4.54PIVPTVDD143 pKa = 3.2TPDD146 pKa = 3.37AQTTLTTNTEE156 pKa = 3.95DD157 pKa = 3.41PAATVSGTGEE167 pKa = 4.0PGSTVEE173 pKa = 5.4VVLGDD178 pKa = 3.48QTLEE182 pKa = 4.02TTIDD186 pKa = 3.67EE187 pKa = 4.87DD188 pKa = 4.39GTWSVTFEE196 pKa = 5.27DD197 pKa = 4.46DD198 pKa = 3.67TFPSDD203 pKa = 3.8GDD205 pKa = 3.68LTSTVDD211 pKa = 3.12VTAPDD216 pKa = 3.61GTEE219 pKa = 3.91YY220 pKa = 10.86TLDD223 pKa = 4.02GPDD226 pKa = 4.19FLIDD230 pKa = 3.52MTPPAVVITEE240 pKa = 4.54GALSTGDD247 pKa = 3.51VEE249 pKa = 5.62NLSEE253 pKa = 4.25YY254 pKa = 10.56QDD256 pKa = 4.35GITVRR261 pKa = 11.84GTGEE265 pKa = 3.61AGAAIVVAVAGHH277 pKa = 6.07AQATTVAADD286 pKa = 4.21GSWSVTFTTEE296 pKa = 3.57QMPAGTYY303 pKa = 9.87SEE305 pKa = 4.68AMTVTATDD313 pKa = 3.11AVGNVTTLNDD323 pKa = 3.66TLVVDD328 pKa = 4.8TEE330 pKa = 4.25ASVAFSSASVTGDD343 pKa = 3.36NIVNADD349 pKa = 3.56EE350 pKa = 4.54AQTGFALSGTSEE362 pKa = 4.25PGSVSVRR369 pKa = 11.84IEE371 pKa = 3.94LGGTSYY377 pKa = 11.36SATPAADD384 pKa = 3.93GTWSIALQDD393 pKa = 3.54VALATGVHH401 pKa = 5.79TATVTATDD409 pKa = 3.81ANGNTASATRR419 pKa = 11.84AISFDD424 pKa = 3.32TDD426 pKa = 3.29TSVSFDD432 pKa = 3.19SAQAGGNNLLNAAEE446 pKa = 4.14RR447 pKa = 11.84SAGLVLTGTAEE458 pKa = 4.07PGATVVVAFEE468 pKa = 4.34QGSHH472 pKa = 4.92TVTAGADD479 pKa = 3.91GHH481 pKa = 6.36WSASFAASEE490 pKa = 4.34VRR492 pKa = 11.84TGTYY496 pKa = 10.45SSTATVTATDD506 pKa = 3.11IAGNVATASHH516 pKa = 7.51TINVDD521 pKa = 3.4TEE523 pKa = 4.52VQNFTHH529 pKa = 6.99TSPTANAIVADD540 pKa = 4.75DD541 pKa = 3.47IVNARR546 pKa = 11.84EE547 pKa = 3.83AAGGLVVTGTVEE559 pKa = 3.84PGATVTVQLASGSVIPAINLGNGSWTATIPAAQLPAVEE597 pKa = 4.54TDD599 pKa = 3.1NVTLTVRR606 pKa = 11.84ATDD609 pKa = 3.59VNGNTATQTSSIDD622 pKa = 3.34FDD624 pKa = 3.78PVVRR628 pKa = 11.84NFAPNATVAGDD639 pKa = 4.26GIVNAQEE646 pKa = 4.19GADD649 pKa = 3.53GFVISGAVEE658 pKa = 3.87PGSRR662 pKa = 11.84VAVTLASGAVEE673 pKa = 4.2TVTAGANGQWSVMFHH688 pKa = 6.72SDD690 pKa = 4.08DD691 pKa = 4.7LGTTGSGTMGYY702 pKa = 9.85QIAATDD708 pKa = 3.8PVGNSATASGSFGYY722 pKa = 10.7DD723 pKa = 2.91LVAPEE728 pKa = 4.93SPDD731 pKa = 3.12IIAFTRR737 pKa = 11.84DD738 pKa = 3.03AGSLLGIRR746 pKa = 11.84TDD748 pKa = 3.53LGEE751 pKa = 4.04NTYY754 pKa = 10.76EE755 pKa = 3.96IAAVDD760 pKa = 3.19AAGRR764 pKa = 11.84VTGVPNDD771 pKa = 3.84ASVSTHH777 pKa = 5.74GGYY780 pKa = 8.1GTYY783 pKa = 10.62DD784 pKa = 4.24FYY786 pKa = 11.99SSVPNGSYY794 pKa = 10.65LVVSDD799 pKa = 4.79HH800 pKa = 6.74DD801 pKa = 3.89TAGNEE806 pKa = 4.0TSTLLVVDD814 pKa = 3.68NTSAVTVDD822 pKa = 4.12LARR825 pKa = 11.84SGLGDD830 pKa = 3.83FDD832 pKa = 5.64FGTIDD837 pKa = 3.47LTFAPEE843 pKa = 3.78AQLTITEE850 pKa = 4.21AQLVALTGPDD860 pKa = 3.35HH861 pKa = 7.0ALVIQGDD868 pKa = 4.28SADD871 pKa = 3.76HH872 pKa = 5.09VTALGAEE879 pKa = 4.22DD880 pKa = 4.16SGQNTVIGGHH890 pKa = 5.67TYY892 pKa = 10.59SVYY895 pKa = 10.76TLGDD899 pKa = 3.87DD900 pKa = 3.97GASLIIDD907 pKa = 3.84DD908 pKa = 5.07QITHH912 pKa = 6.02LTII915 pKa = 4.16
Molecular weight: 91.8 kDa
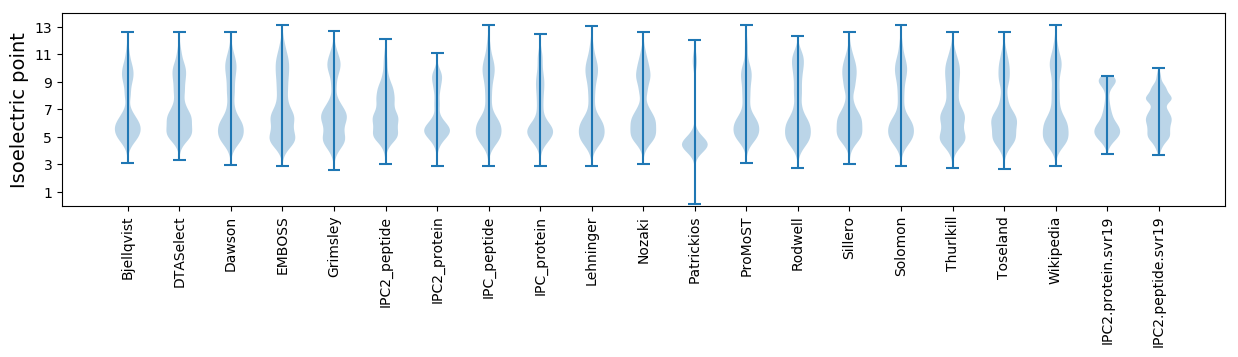
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T4JK15|A0A2T4JK15_9RHOB DUF3429 domain-containing protein OS=Rhodobacter veldkampii DSM 11550 OX=1185920 GN=C5F46_05720 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.12GGRR28 pKa = 11.84RR29 pKa = 11.84VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.0GRR39 pKa = 11.84KK40 pKa = 8.91SLSAA44 pKa = 3.86
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.12GGRR28 pKa = 11.84RR29 pKa = 11.84VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.0GRR39 pKa = 11.84KK40 pKa = 8.91SLSAA44 pKa = 3.86
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
961833 |
28 |
2468 |
308.8 |
33.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.249 ± 0.084 | 0.857 ± 0.014 |
5.696 ± 0.032 | 5.571 ± 0.043 |
3.509 ± 0.026 | 8.877 ± 0.046 |
1.999 ± 0.021 | 4.742 ± 0.033 |
2.744 ± 0.041 | 10.447 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.61 ± 0.019 | 2.184 ± 0.02 |
5.494 ± 0.036 | 3.021 ± 0.021 |
7.6 ± 0.043 | 4.446 ± 0.027 |
5.243 ± 0.033 | 7.269 ± 0.036 |
1.437 ± 0.022 | 2.005 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
