
Hubei myriapoda virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
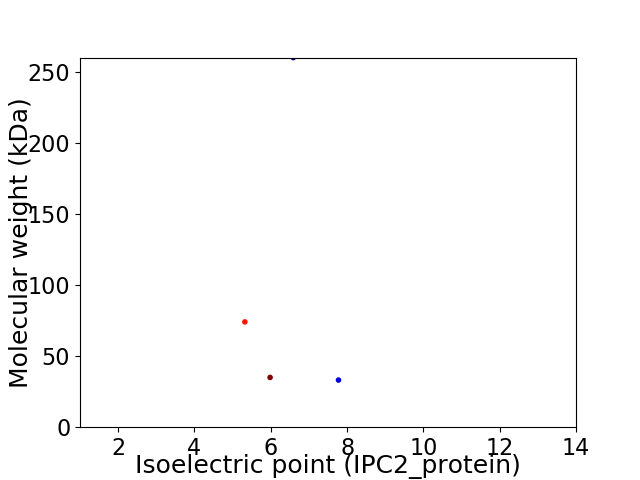
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJG9|A0A1L3KJG9_9VIRU Uncharacterized protein OS=Hubei myriapoda virus 3 OX=1922932 PE=4 SV=1
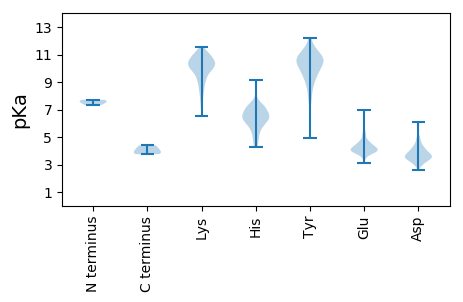
MM1 pKa = 7.33SKK3 pKa = 8.4EE4 pKa = 4.23TKK6 pKa = 10.59AEE8 pKa = 3.85AVPEE12 pKa = 4.24NNPSLAFTNEE22 pKa = 4.06DD23 pKa = 2.98VSLPTSHH30 pKa = 7.22SAFTEE35 pKa = 4.53PGTSPSTVARR45 pKa = 11.84DD46 pKa = 3.45IGCTGVRR53 pKa = 11.84VFPEE57 pKa = 4.91LYY59 pKa = 10.27EE60 pKa = 4.7KK61 pKa = 10.46DD62 pKa = 3.87YY63 pKa = 11.36QIDD66 pKa = 4.12SITWDD71 pKa = 3.04IGKK74 pKa = 9.72SPGTILKK81 pKa = 9.97VFTISPDD88 pKa = 3.14MHH90 pKa = 6.29PQIAYY95 pKa = 9.68LMKK98 pKa = 9.99MFQFWSGTIDD108 pKa = 3.05IVFKK112 pKa = 10.84IAGTGFNAGLIGITTANPLVPLEE135 pKa = 4.39TYY137 pKa = 11.11KK138 pKa = 10.05NTTDD142 pKa = 3.68FCNFDD147 pKa = 3.37TQYY150 pKa = 11.93ADD152 pKa = 3.7PKK154 pKa = 10.52QLALFKK160 pKa = 9.67VTPQDD165 pKa = 2.96FHH167 pKa = 9.14IYY169 pKa = 7.71NTHH172 pKa = 6.3INSINQTATGDD183 pKa = 3.41TDD185 pKa = 3.47ADD187 pKa = 4.37KK188 pKa = 10.05ITKK191 pKa = 10.33ASGLAAKK198 pKa = 10.14VAVFVSLQLASGTSGNTSVDD218 pKa = 2.58IGVFLVASKK227 pKa = 10.58SFRR230 pKa = 11.84LHH232 pKa = 6.27QILPAPSSITNNISFEE248 pKa = 4.41LLNHH252 pKa = 6.57ALDD255 pKa = 3.58VSNPYY260 pKa = 9.69TKK262 pKa = 9.9TATLEE267 pKa = 3.87RR268 pKa = 11.84CRR270 pKa = 11.84GMMAVPLSLLDD281 pKa = 3.45KK282 pKa = 10.01PKK284 pKa = 10.9RR285 pKa = 11.84FIANCAGATGWEE297 pKa = 3.83NDD299 pKa = 2.85RR300 pKa = 11.84RR301 pKa = 11.84IWNKK305 pKa = 6.52THH307 pKa = 7.11TYY309 pKa = 10.77NSINGPIVIEE319 pKa = 4.16NVTYY323 pKa = 10.17DD324 pKa = 3.5YY325 pKa = 11.06PAFLGFYY332 pKa = 8.81EE333 pKa = 4.27SHH335 pKa = 6.73PTTVVSIPVNQNDD348 pKa = 3.48EE349 pKa = 4.5TALSSHH355 pKa = 6.17VATLDD360 pKa = 3.27YY361 pKa = 10.7TDD363 pKa = 4.1YY364 pKa = 11.24FEE366 pKa = 4.5EE367 pKa = 5.15HH368 pKa = 6.96PEE370 pKa = 3.97EE371 pKa = 4.68KK372 pKa = 10.3FVGVGFYY379 pKa = 10.05TKK381 pKa = 10.69NCIPASLIPRR391 pKa = 11.84KK392 pKa = 9.79SVLQNIRR399 pKa = 11.84TVEE402 pKa = 4.06SQNLDD407 pKa = 3.35GALGLYY413 pKa = 10.57AIDD416 pKa = 4.99NPEE419 pKa = 3.98EE420 pKa = 4.12YY421 pKa = 10.56QNLTSMLPQHH431 pKa = 6.48PRR433 pKa = 11.84FDD435 pKa = 4.23SIRR438 pKa = 11.84NSFQWGLAMINPTTKK453 pKa = 10.27HH454 pKa = 4.56YY455 pKa = 9.67TNLHH459 pKa = 5.68SFTTLKK465 pKa = 10.81NLTCTPPDD473 pKa = 3.42WEE475 pKa = 5.31LDD477 pKa = 3.76FSLKK481 pKa = 10.49GPSSGEE487 pKa = 3.95SFLLFTDD494 pKa = 4.28TYY496 pKa = 10.91DD497 pKa = 3.48YY498 pKa = 11.25QAADD502 pKa = 5.29LISIDD507 pKa = 3.08IGLPSFQTEE516 pKa = 3.91HH517 pKa = 6.7LASYY521 pKa = 8.78MYY523 pKa = 10.6NKK525 pKa = 9.56PIQSNQSVLLSMVDD539 pKa = 3.62TIDD542 pKa = 4.71DD543 pKa = 4.7KK544 pKa = 11.32IVSQAKK550 pKa = 9.87LYY552 pKa = 9.05PEE554 pKa = 5.25GYY556 pKa = 7.69ITVPTVSQLTFYY568 pKa = 11.22GIDD571 pKa = 3.46RR572 pKa = 11.84YY573 pKa = 11.04KK574 pKa = 11.01LIAQRR579 pKa = 11.84VVPRR583 pKa = 11.84NYY585 pKa = 10.3EE586 pKa = 3.74MRR588 pKa = 11.84PTAVQNLVTNMIAARR603 pKa = 11.84TQQEE607 pKa = 4.31SQTTYY612 pKa = 6.43QTKK615 pKa = 8.43YY616 pKa = 9.34TNFLLSQHH624 pKa = 6.5KK625 pKa = 10.53LLTKK629 pKa = 10.23KK630 pKa = 10.65FKK632 pKa = 10.35TVDD635 pKa = 3.4KK636 pKa = 10.59LNKK639 pKa = 10.12YY640 pKa = 10.32LDD642 pKa = 3.58EE643 pKa = 5.87HH644 pKa = 5.05STKK647 pKa = 9.93WPAEE651 pKa = 3.98PLPPEE656 pKa = 4.31QPSAPDD662 pKa = 3.13SS663 pKa = 3.75
MM1 pKa = 7.33SKK3 pKa = 8.4EE4 pKa = 4.23TKK6 pKa = 10.59AEE8 pKa = 3.85AVPEE12 pKa = 4.24NNPSLAFTNEE22 pKa = 4.06DD23 pKa = 2.98VSLPTSHH30 pKa = 7.22SAFTEE35 pKa = 4.53PGTSPSTVARR45 pKa = 11.84DD46 pKa = 3.45IGCTGVRR53 pKa = 11.84VFPEE57 pKa = 4.91LYY59 pKa = 10.27EE60 pKa = 4.7KK61 pKa = 10.46DD62 pKa = 3.87YY63 pKa = 11.36QIDD66 pKa = 4.12SITWDD71 pKa = 3.04IGKK74 pKa = 9.72SPGTILKK81 pKa = 9.97VFTISPDD88 pKa = 3.14MHH90 pKa = 6.29PQIAYY95 pKa = 9.68LMKK98 pKa = 9.99MFQFWSGTIDD108 pKa = 3.05IVFKK112 pKa = 10.84IAGTGFNAGLIGITTANPLVPLEE135 pKa = 4.39TYY137 pKa = 11.11KK138 pKa = 10.05NTTDD142 pKa = 3.68FCNFDD147 pKa = 3.37TQYY150 pKa = 11.93ADD152 pKa = 3.7PKK154 pKa = 10.52QLALFKK160 pKa = 9.67VTPQDD165 pKa = 2.96FHH167 pKa = 9.14IYY169 pKa = 7.71NTHH172 pKa = 6.3INSINQTATGDD183 pKa = 3.41TDD185 pKa = 3.47ADD187 pKa = 4.37KK188 pKa = 10.05ITKK191 pKa = 10.33ASGLAAKK198 pKa = 10.14VAVFVSLQLASGTSGNTSVDD218 pKa = 2.58IGVFLVASKK227 pKa = 10.58SFRR230 pKa = 11.84LHH232 pKa = 6.27QILPAPSSITNNISFEE248 pKa = 4.41LLNHH252 pKa = 6.57ALDD255 pKa = 3.58VSNPYY260 pKa = 9.69TKK262 pKa = 9.9TATLEE267 pKa = 3.87RR268 pKa = 11.84CRR270 pKa = 11.84GMMAVPLSLLDD281 pKa = 3.45KK282 pKa = 10.01PKK284 pKa = 10.9RR285 pKa = 11.84FIANCAGATGWEE297 pKa = 3.83NDD299 pKa = 2.85RR300 pKa = 11.84RR301 pKa = 11.84IWNKK305 pKa = 6.52THH307 pKa = 7.11TYY309 pKa = 10.77NSINGPIVIEE319 pKa = 4.16NVTYY323 pKa = 10.17DD324 pKa = 3.5YY325 pKa = 11.06PAFLGFYY332 pKa = 8.81EE333 pKa = 4.27SHH335 pKa = 6.73PTTVVSIPVNQNDD348 pKa = 3.48EE349 pKa = 4.5TALSSHH355 pKa = 6.17VATLDD360 pKa = 3.27YY361 pKa = 10.7TDD363 pKa = 4.1YY364 pKa = 11.24FEE366 pKa = 4.5EE367 pKa = 5.15HH368 pKa = 6.96PEE370 pKa = 3.97EE371 pKa = 4.68KK372 pKa = 10.3FVGVGFYY379 pKa = 10.05TKK381 pKa = 10.69NCIPASLIPRR391 pKa = 11.84KK392 pKa = 9.79SVLQNIRR399 pKa = 11.84TVEE402 pKa = 4.06SQNLDD407 pKa = 3.35GALGLYY413 pKa = 10.57AIDD416 pKa = 4.99NPEE419 pKa = 3.98EE420 pKa = 4.12YY421 pKa = 10.56QNLTSMLPQHH431 pKa = 6.48PRR433 pKa = 11.84FDD435 pKa = 4.23SIRR438 pKa = 11.84NSFQWGLAMINPTTKK453 pKa = 10.27HH454 pKa = 4.56YY455 pKa = 9.67TNLHH459 pKa = 5.68SFTTLKK465 pKa = 10.81NLTCTPPDD473 pKa = 3.42WEE475 pKa = 5.31LDD477 pKa = 3.76FSLKK481 pKa = 10.49GPSSGEE487 pKa = 3.95SFLLFTDD494 pKa = 4.28TYY496 pKa = 10.91DD497 pKa = 3.48YY498 pKa = 11.25QAADD502 pKa = 5.29LISIDD507 pKa = 3.08IGLPSFQTEE516 pKa = 3.91HH517 pKa = 6.7LASYY521 pKa = 8.78MYY523 pKa = 10.6NKK525 pKa = 9.56PIQSNQSVLLSMVDD539 pKa = 3.62TIDD542 pKa = 4.71DD543 pKa = 4.7KK544 pKa = 11.32IVSQAKK550 pKa = 9.87LYY552 pKa = 9.05PEE554 pKa = 5.25GYY556 pKa = 7.69ITVPTVSQLTFYY568 pKa = 11.22GIDD571 pKa = 3.46RR572 pKa = 11.84YY573 pKa = 11.04KK574 pKa = 11.01LIAQRR579 pKa = 11.84VVPRR583 pKa = 11.84NYY585 pKa = 10.3EE586 pKa = 3.74MRR588 pKa = 11.84PTAVQNLVTNMIAARR603 pKa = 11.84TQQEE607 pKa = 4.31SQTTYY612 pKa = 6.43QTKK615 pKa = 8.43YY616 pKa = 9.34TNFLLSQHH624 pKa = 6.5KK625 pKa = 10.53LLTKK629 pKa = 10.23KK630 pKa = 10.65FKK632 pKa = 10.35TVDD635 pKa = 3.4KK636 pKa = 10.59LNKK639 pKa = 10.12YY640 pKa = 10.32LDD642 pKa = 3.58EE643 pKa = 5.87HH644 pKa = 5.05STKK647 pKa = 9.93WPAEE651 pKa = 3.98PLPPEE656 pKa = 4.31QPSAPDD662 pKa = 3.13SS663 pKa = 3.75
Molecular weight: 74.03 kDa
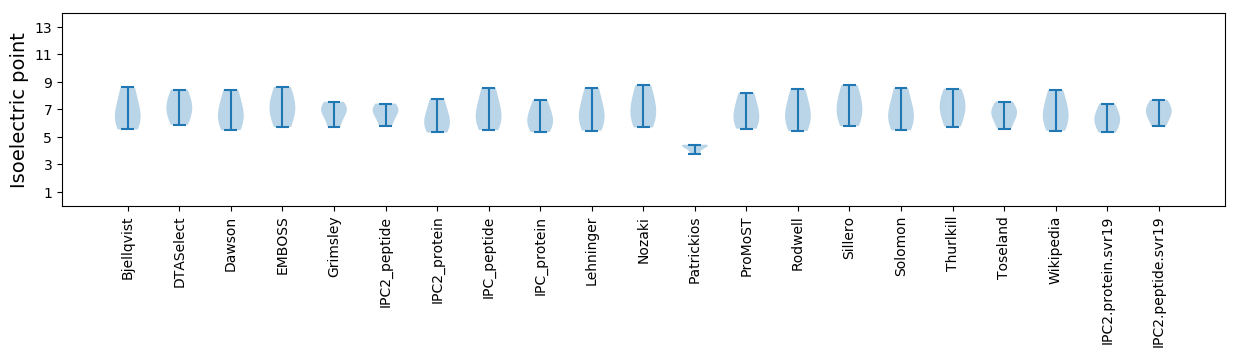
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJL3|A0A1L3KJL3_9VIRU Uncharacterized protein OS=Hubei myriapoda virus 3 OX=1922932 PE=4 SV=1
MM1 pKa = 7.62PFLYY5 pKa = 10.24KK6 pKa = 9.9IEE8 pKa = 4.47SLIQLLDD15 pKa = 4.36FIEE18 pKa = 5.12DD19 pKa = 3.42QFQVDD24 pKa = 4.53FEE26 pKa = 5.04LGNSIYY32 pKa = 11.01YY33 pKa = 10.6LLQFPKK39 pKa = 9.78PWKK42 pKa = 9.26PHH44 pKa = 4.85TNEE47 pKa = 3.38PWFLYY52 pKa = 10.31YY53 pKa = 10.51CDD55 pKa = 3.7IYY57 pKa = 11.32EE58 pKa = 4.19HH59 pKa = 6.65FKK61 pKa = 10.32RR62 pKa = 11.84TIEE65 pKa = 4.09NDD67 pKa = 3.59PQHH70 pKa = 6.44LHH72 pKa = 7.15PDD74 pKa = 3.53LGVTIDD80 pKa = 4.33FLPDD84 pKa = 3.38PEE86 pKa = 4.71SFVAGATNSKK96 pKa = 10.41LFVYY100 pKa = 10.0KK101 pKa = 9.97YY102 pKa = 10.52QKK104 pKa = 10.53NPRR107 pKa = 11.84KK108 pKa = 8.88KK109 pKa = 8.72TNLLAHH115 pKa = 6.61TLNCVCEE122 pKa = 3.98DD123 pKa = 3.68CKK125 pKa = 11.21YY126 pKa = 10.97SEE128 pKa = 4.19QLALALDD135 pKa = 3.87RR136 pKa = 11.84KK137 pKa = 10.39DD138 pKa = 3.91RR139 pKa = 11.84LVSAIKK145 pKa = 10.44RR146 pKa = 11.84KK147 pKa = 8.04TDD149 pKa = 3.16HH150 pKa = 6.85NGQLLQRR157 pKa = 11.84PKK159 pKa = 10.59FGQKK163 pKa = 10.18KK164 pKa = 6.95IQNYY168 pKa = 8.13IRR170 pKa = 11.84NSGSEE175 pKa = 3.9DD176 pKa = 3.6LFRR179 pKa = 11.84QHH181 pKa = 6.81TSGCSANQLYY191 pKa = 10.85SSLAKK196 pKa = 8.57YY197 pKa = 7.31TKK199 pKa = 8.48PTVEE203 pKa = 3.86ITSPTRR209 pKa = 11.84DD210 pKa = 3.23MLDD213 pKa = 3.31FRR215 pKa = 11.84FKK217 pKa = 10.15EE218 pKa = 4.07VQRR221 pKa = 11.84KK222 pKa = 9.1KK223 pKa = 9.8EE224 pKa = 4.12TLSDD228 pKa = 3.61EE229 pKa = 5.3LISKK233 pKa = 9.25LAIALTSTGLPDD245 pKa = 5.91AIPKK249 pKa = 9.85ILRR252 pKa = 11.84QLKK255 pKa = 9.46VPEE258 pKa = 4.26GKK260 pKa = 9.06IDD262 pKa = 3.38QLAYY266 pKa = 10.61DD267 pKa = 4.36NQTVPPDD274 pKa = 3.49VTPKK278 pKa = 10.89NKK280 pKa = 8.67TQSLPLL286 pKa = 4.39
MM1 pKa = 7.62PFLYY5 pKa = 10.24KK6 pKa = 9.9IEE8 pKa = 4.47SLIQLLDD15 pKa = 4.36FIEE18 pKa = 5.12DD19 pKa = 3.42QFQVDD24 pKa = 4.53FEE26 pKa = 5.04LGNSIYY32 pKa = 11.01YY33 pKa = 10.6LLQFPKK39 pKa = 9.78PWKK42 pKa = 9.26PHH44 pKa = 4.85TNEE47 pKa = 3.38PWFLYY52 pKa = 10.31YY53 pKa = 10.51CDD55 pKa = 3.7IYY57 pKa = 11.32EE58 pKa = 4.19HH59 pKa = 6.65FKK61 pKa = 10.32RR62 pKa = 11.84TIEE65 pKa = 4.09NDD67 pKa = 3.59PQHH70 pKa = 6.44LHH72 pKa = 7.15PDD74 pKa = 3.53LGVTIDD80 pKa = 4.33FLPDD84 pKa = 3.38PEE86 pKa = 4.71SFVAGATNSKK96 pKa = 10.41LFVYY100 pKa = 10.0KK101 pKa = 9.97YY102 pKa = 10.52QKK104 pKa = 10.53NPRR107 pKa = 11.84KK108 pKa = 8.88KK109 pKa = 8.72TNLLAHH115 pKa = 6.61TLNCVCEE122 pKa = 3.98DD123 pKa = 3.68CKK125 pKa = 11.21YY126 pKa = 10.97SEE128 pKa = 4.19QLALALDD135 pKa = 3.87RR136 pKa = 11.84KK137 pKa = 10.39DD138 pKa = 3.91RR139 pKa = 11.84LVSAIKK145 pKa = 10.44RR146 pKa = 11.84KK147 pKa = 8.04TDD149 pKa = 3.16HH150 pKa = 6.85NGQLLQRR157 pKa = 11.84PKK159 pKa = 10.59FGQKK163 pKa = 10.18KK164 pKa = 6.95IQNYY168 pKa = 8.13IRR170 pKa = 11.84NSGSEE175 pKa = 3.9DD176 pKa = 3.6LFRR179 pKa = 11.84QHH181 pKa = 6.81TSGCSANQLYY191 pKa = 10.85SSLAKK196 pKa = 8.57YY197 pKa = 7.31TKK199 pKa = 8.48PTVEE203 pKa = 3.86ITSPTRR209 pKa = 11.84DD210 pKa = 3.23MLDD213 pKa = 3.31FRR215 pKa = 11.84FKK217 pKa = 10.15EE218 pKa = 4.07VQRR221 pKa = 11.84KK222 pKa = 9.1KK223 pKa = 9.8EE224 pKa = 4.12TLSDD228 pKa = 3.61EE229 pKa = 5.3LISKK233 pKa = 9.25LAIALTSTGLPDD245 pKa = 5.91AIPKK249 pKa = 9.85ILRR252 pKa = 11.84QLKK255 pKa = 9.46VPEE258 pKa = 4.26GKK260 pKa = 9.06IDD262 pKa = 3.38QLAYY266 pKa = 10.61DD267 pKa = 4.36NQTVPPDD274 pKa = 3.49VTPKK278 pKa = 10.89NKK280 pKa = 8.67TQSLPLL286 pKa = 4.39
Molecular weight: 33.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3510 |
286 |
2242 |
877.5 |
100.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.014 ± 0.761 | 1.51 ± 0.403 |
6.296 ± 0.326 | 5.755 ± 0.451 |
5.726 ± 0.707 | 4.53 ± 0.483 |
2.735 ± 0.12 | 6.838 ± 0.522 |
7.664 ± 0.803 | 9.516 ± 0.636 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.909 ± 0.196 | 5.641 ± 0.302 |
5.071 ± 1.01 | 4.957 ± 0.3 |
3.903 ± 0.472 | 6.781 ± 0.884 |
6.581 ± 1.083 | 4.815 ± 0.272 |
0.969 ± 0.044 | 3.789 ± 0.227 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
