
Beihai narna-like virus 11
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
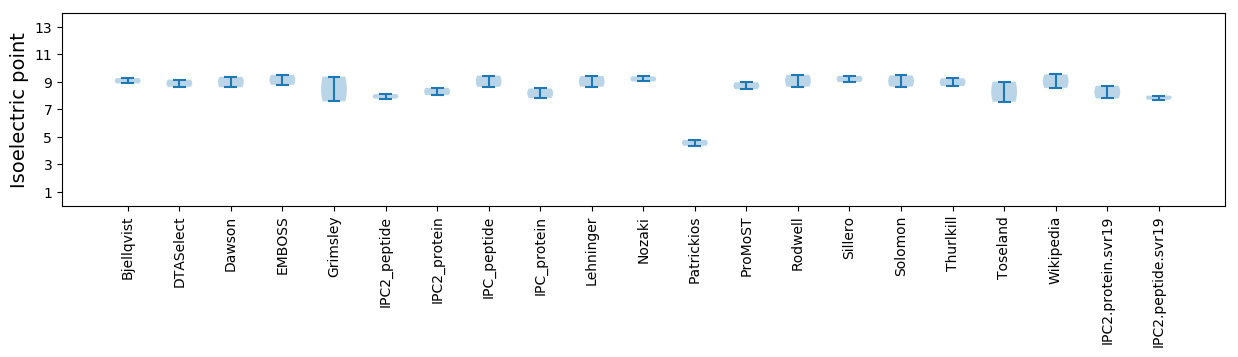
Average proteome isoelectric point is 8.26
Get precalculated fractions of proteins
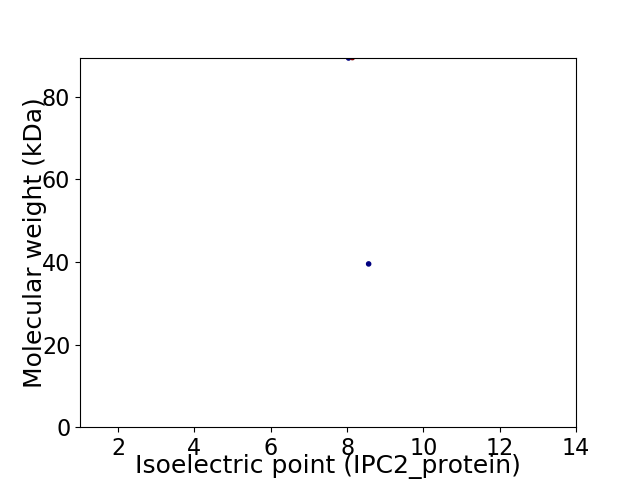
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KI60|A0A1L3KI60_9VIRU Uncharacterized protein OS=Beihai narna-like virus 11 OX=1922438 PE=4 SV=1
MM1 pKa = 7.52ASDD4 pKa = 3.41HH5 pKa = 6.22RR6 pKa = 11.84TDD8 pKa = 3.1KK9 pKa = 11.09GDD11 pKa = 3.34RR12 pKa = 11.84VEE14 pKa = 4.34AKK16 pKa = 9.65TYY18 pKa = 10.84LSLIDD23 pKa = 4.93VISQSLRR30 pKa = 11.84PSGDD34 pKa = 2.98GKK36 pKa = 10.91ASSCCRR42 pKa = 11.84CYY44 pKa = 10.25EE45 pKa = 4.12ASSLACQLIRR55 pKa = 11.84RR56 pKa = 11.84HH57 pKa = 5.9SGVLGSAVVKK67 pKa = 10.42KK68 pKa = 10.23RR69 pKa = 11.84SKK71 pKa = 10.48GKK73 pKa = 10.16EE74 pKa = 3.79VIQWPDD80 pKa = 3.62FPQGDD85 pKa = 4.39VVSSLYY91 pKa = 8.8FTRR94 pKa = 11.84SYY96 pKa = 11.52LEE98 pKa = 3.75VLIDD102 pKa = 4.25FDD104 pKa = 6.09DD105 pKa = 6.38RR106 pKa = 11.84LLKK109 pKa = 10.26WNWKK113 pKa = 7.67HH114 pKa = 6.22LSVIFEE120 pKa = 4.49EE121 pKa = 4.69CMSWEE126 pKa = 3.71PLNFIKK132 pKa = 10.45HH133 pKa = 6.11AKK135 pKa = 9.34FCTAVPMADD144 pKa = 3.36YY145 pKa = 11.3LNRR148 pKa = 11.84KK149 pKa = 8.38LKK151 pKa = 10.36NPRR154 pKa = 11.84PALVEE159 pKa = 3.69RR160 pKa = 11.84VGPYY164 pKa = 9.98FRR166 pKa = 11.84GPLKK170 pKa = 10.45RR171 pKa = 11.84YY172 pKa = 6.32LTNRR176 pKa = 11.84INTGRR181 pKa = 11.84GKK183 pKa = 10.04NLSLAWSILQGVKK196 pKa = 10.24RR197 pKa = 11.84GCAPASDD204 pKa = 4.87EE205 pKa = 4.2YY206 pKa = 11.38LQSEE210 pKa = 4.74YY211 pKa = 11.28NSYY214 pKa = 11.68ADD216 pKa = 5.23LLGEE220 pKa = 4.47PPSNPYY226 pKa = 10.48SDD228 pKa = 3.4YY229 pKa = 9.8TRR231 pKa = 11.84YY232 pKa = 10.3YY233 pKa = 10.37SEE235 pKa = 3.57IWDD238 pKa = 3.63GLRR241 pKa = 11.84VRR243 pKa = 11.84RR244 pKa = 11.84EE245 pKa = 3.61THH247 pKa = 5.42EE248 pKa = 4.51FYY250 pKa = 10.31PSKK253 pKa = 10.35SAKK256 pKa = 10.3LGFNRR261 pKa = 11.84SEE263 pKa = 4.17GGGVQGMVRR272 pKa = 11.84SMDD275 pKa = 3.69LEE277 pKa = 5.73AGLVDD282 pKa = 5.42MVDD285 pKa = 3.23TDD287 pKa = 3.11KK288 pKa = 11.67GVIEE292 pKa = 4.02RR293 pKa = 11.84RR294 pKa = 11.84GVNIPPLSDD303 pKa = 3.5LLTAEE308 pKa = 4.39SEE310 pKa = 4.47EE311 pKa = 4.17PCIVRR316 pKa = 11.84VQGLQEE322 pKa = 3.8PLKK325 pKa = 10.87VRR327 pKa = 11.84FITKK331 pKa = 9.43SPSVRR336 pKa = 11.84SYY338 pKa = 10.21VCKK341 pKa = 10.24PIQNALWNHH350 pKa = 5.85LKK352 pKa = 10.42EE353 pKa = 4.45SEE355 pKa = 4.05PFALIGHH362 pKa = 6.82PIRR365 pKa = 11.84RR366 pKa = 11.84EE367 pKa = 3.73DD368 pKa = 4.22LRR370 pKa = 11.84QLKK373 pKa = 10.0IKK375 pKa = 9.49SLRR378 pKa = 11.84FDD380 pKa = 4.71FGPQPFWVSGDD391 pKa = 3.67YY392 pKa = 10.74KK393 pKa = 10.92SATDD397 pKa = 5.5RR398 pKa = 11.84IDD400 pKa = 3.73LRR402 pKa = 11.84QTKK405 pKa = 10.11DD406 pKa = 3.24ALEE409 pKa = 4.34CALAEE414 pKa = 4.26LLLHH418 pKa = 5.63GHH420 pKa = 6.41IGLRR424 pKa = 11.84QADD427 pKa = 3.39ACRR430 pKa = 11.84RR431 pKa = 11.84EE432 pKa = 4.84LYY434 pKa = 9.91EE435 pKa = 3.82QTITFPLHH443 pKa = 5.85LQMPSVPQGNGQLMGSVLSFPILCVINLAAYY474 pKa = 6.84WSSMEE479 pKa = 4.58EE480 pKa = 4.2YY481 pKa = 10.11FQEE484 pKa = 4.56EE485 pKa = 4.76YY486 pKa = 10.69RR487 pKa = 11.84LQDD490 pKa = 3.64LPVLINGDD498 pKa = 3.96DD499 pKa = 3.9IGFQSNDD506 pKa = 2.45EE507 pKa = 5.05HH508 pKa = 7.37YY509 pKa = 10.86AIWQQKK515 pKa = 6.23IHH517 pKa = 6.14EE518 pKa = 4.65VGLKK522 pKa = 10.45LSIGKK527 pKa = 9.89NYY529 pKa = 9.69IHH531 pKa = 6.78PTYY534 pKa = 9.78FTINSEE540 pKa = 4.09LWNMCDD546 pKa = 3.0LTQVHH551 pKa = 6.67FFNLGCLTNMNRR563 pKa = 11.84RR564 pKa = 11.84TGRR567 pKa = 11.84EE568 pKa = 4.1CTKK571 pKa = 10.45SLPIAEE577 pKa = 4.81LWSEE581 pKa = 4.21VHH583 pKa = 6.84VGATNKK589 pKa = 9.82DD590 pKa = 3.56RR591 pKa = 11.84AWRR594 pKa = 11.84RR595 pKa = 11.84FVHH598 pKa = 5.69YY599 pKa = 9.62NKK601 pKa = 10.04PQIQSATLRR610 pKa = 11.84GRR612 pKa = 11.84LNLGLPYY619 pKa = 9.97TAGGLNCSIPCDD631 pKa = 3.67YY632 pKa = 10.68RR633 pKa = 11.84VTDD636 pKa = 3.78GQRR639 pKa = 11.84RR640 pKa = 11.84LQRR643 pKa = 11.84YY644 pKa = 8.03CEE646 pKa = 3.47RR647 pKa = 11.84LYY649 pKa = 11.03RR650 pKa = 11.84NEE652 pKa = 3.31GRR654 pKa = 11.84TLTGVKK660 pKa = 9.92GRR662 pKa = 11.84SHH664 pKa = 6.71MLHH667 pKa = 5.32KK668 pKa = 10.27HH669 pKa = 4.54GRR671 pKa = 11.84SNNMRR676 pKa = 11.84LYY678 pKa = 10.5DD679 pKa = 3.51GTYY682 pKa = 10.29EE683 pKa = 4.35VILSSSEE690 pKa = 3.82GQEE693 pKa = 3.34QRR695 pKa = 11.84VPVEE699 pKa = 4.23LLDD702 pKa = 4.35APRR705 pKa = 11.84EE706 pKa = 4.12DD707 pKa = 3.72IILPVKK713 pKa = 10.48IPTEE717 pKa = 3.97THH719 pKa = 6.72LSPFGPILADD729 pKa = 3.82DD730 pKa = 3.77EE731 pKa = 4.91VEE733 pKa = 4.5NKK735 pKa = 9.91FSYY738 pKa = 10.27PSRR741 pKa = 11.84KK742 pKa = 10.31DD743 pKa = 2.89FDD745 pKa = 3.75QAKK748 pKa = 9.44KK749 pKa = 9.53FAPGGSPKK757 pKa = 10.38NFCLTKK763 pKa = 10.26KK764 pKa = 10.35VPYY767 pKa = 9.87DD768 pKa = 3.3KK769 pKa = 10.99RR770 pKa = 11.84FVTGKK775 pKa = 9.74VIKK778 pKa = 8.6VTPYY782 pKa = 10.67
MM1 pKa = 7.52ASDD4 pKa = 3.41HH5 pKa = 6.22RR6 pKa = 11.84TDD8 pKa = 3.1KK9 pKa = 11.09GDD11 pKa = 3.34RR12 pKa = 11.84VEE14 pKa = 4.34AKK16 pKa = 9.65TYY18 pKa = 10.84LSLIDD23 pKa = 4.93VISQSLRR30 pKa = 11.84PSGDD34 pKa = 2.98GKK36 pKa = 10.91ASSCCRR42 pKa = 11.84CYY44 pKa = 10.25EE45 pKa = 4.12ASSLACQLIRR55 pKa = 11.84RR56 pKa = 11.84HH57 pKa = 5.9SGVLGSAVVKK67 pKa = 10.42KK68 pKa = 10.23RR69 pKa = 11.84SKK71 pKa = 10.48GKK73 pKa = 10.16EE74 pKa = 3.79VIQWPDD80 pKa = 3.62FPQGDD85 pKa = 4.39VVSSLYY91 pKa = 8.8FTRR94 pKa = 11.84SYY96 pKa = 11.52LEE98 pKa = 3.75VLIDD102 pKa = 4.25FDD104 pKa = 6.09DD105 pKa = 6.38RR106 pKa = 11.84LLKK109 pKa = 10.26WNWKK113 pKa = 7.67HH114 pKa = 6.22LSVIFEE120 pKa = 4.49EE121 pKa = 4.69CMSWEE126 pKa = 3.71PLNFIKK132 pKa = 10.45HH133 pKa = 6.11AKK135 pKa = 9.34FCTAVPMADD144 pKa = 3.36YY145 pKa = 11.3LNRR148 pKa = 11.84KK149 pKa = 8.38LKK151 pKa = 10.36NPRR154 pKa = 11.84PALVEE159 pKa = 3.69RR160 pKa = 11.84VGPYY164 pKa = 9.98FRR166 pKa = 11.84GPLKK170 pKa = 10.45RR171 pKa = 11.84YY172 pKa = 6.32LTNRR176 pKa = 11.84INTGRR181 pKa = 11.84GKK183 pKa = 10.04NLSLAWSILQGVKK196 pKa = 10.24RR197 pKa = 11.84GCAPASDD204 pKa = 4.87EE205 pKa = 4.2YY206 pKa = 11.38LQSEE210 pKa = 4.74YY211 pKa = 11.28NSYY214 pKa = 11.68ADD216 pKa = 5.23LLGEE220 pKa = 4.47PPSNPYY226 pKa = 10.48SDD228 pKa = 3.4YY229 pKa = 9.8TRR231 pKa = 11.84YY232 pKa = 10.3YY233 pKa = 10.37SEE235 pKa = 3.57IWDD238 pKa = 3.63GLRR241 pKa = 11.84VRR243 pKa = 11.84RR244 pKa = 11.84EE245 pKa = 3.61THH247 pKa = 5.42EE248 pKa = 4.51FYY250 pKa = 10.31PSKK253 pKa = 10.35SAKK256 pKa = 10.3LGFNRR261 pKa = 11.84SEE263 pKa = 4.17GGGVQGMVRR272 pKa = 11.84SMDD275 pKa = 3.69LEE277 pKa = 5.73AGLVDD282 pKa = 5.42MVDD285 pKa = 3.23TDD287 pKa = 3.11KK288 pKa = 11.67GVIEE292 pKa = 4.02RR293 pKa = 11.84RR294 pKa = 11.84GVNIPPLSDD303 pKa = 3.5LLTAEE308 pKa = 4.39SEE310 pKa = 4.47EE311 pKa = 4.17PCIVRR316 pKa = 11.84VQGLQEE322 pKa = 3.8PLKK325 pKa = 10.87VRR327 pKa = 11.84FITKK331 pKa = 9.43SPSVRR336 pKa = 11.84SYY338 pKa = 10.21VCKK341 pKa = 10.24PIQNALWNHH350 pKa = 5.85LKK352 pKa = 10.42EE353 pKa = 4.45SEE355 pKa = 4.05PFALIGHH362 pKa = 6.82PIRR365 pKa = 11.84RR366 pKa = 11.84EE367 pKa = 3.73DD368 pKa = 4.22LRR370 pKa = 11.84QLKK373 pKa = 10.0IKK375 pKa = 9.49SLRR378 pKa = 11.84FDD380 pKa = 4.71FGPQPFWVSGDD391 pKa = 3.67YY392 pKa = 10.74KK393 pKa = 10.92SATDD397 pKa = 5.5RR398 pKa = 11.84IDD400 pKa = 3.73LRR402 pKa = 11.84QTKK405 pKa = 10.11DD406 pKa = 3.24ALEE409 pKa = 4.34CALAEE414 pKa = 4.26LLLHH418 pKa = 5.63GHH420 pKa = 6.41IGLRR424 pKa = 11.84QADD427 pKa = 3.39ACRR430 pKa = 11.84RR431 pKa = 11.84EE432 pKa = 4.84LYY434 pKa = 9.91EE435 pKa = 3.82QTITFPLHH443 pKa = 5.85LQMPSVPQGNGQLMGSVLSFPILCVINLAAYY474 pKa = 6.84WSSMEE479 pKa = 4.58EE480 pKa = 4.2YY481 pKa = 10.11FQEE484 pKa = 4.56EE485 pKa = 4.76YY486 pKa = 10.69RR487 pKa = 11.84LQDD490 pKa = 3.64LPVLINGDD498 pKa = 3.96DD499 pKa = 3.9IGFQSNDD506 pKa = 2.45EE507 pKa = 5.05HH508 pKa = 7.37YY509 pKa = 10.86AIWQQKK515 pKa = 6.23IHH517 pKa = 6.14EE518 pKa = 4.65VGLKK522 pKa = 10.45LSIGKK527 pKa = 9.89NYY529 pKa = 9.69IHH531 pKa = 6.78PTYY534 pKa = 9.78FTINSEE540 pKa = 4.09LWNMCDD546 pKa = 3.0LTQVHH551 pKa = 6.67FFNLGCLTNMNRR563 pKa = 11.84RR564 pKa = 11.84TGRR567 pKa = 11.84EE568 pKa = 4.1CTKK571 pKa = 10.45SLPIAEE577 pKa = 4.81LWSEE581 pKa = 4.21VHH583 pKa = 6.84VGATNKK589 pKa = 9.82DD590 pKa = 3.56RR591 pKa = 11.84AWRR594 pKa = 11.84RR595 pKa = 11.84FVHH598 pKa = 5.69YY599 pKa = 9.62NKK601 pKa = 10.04PQIQSATLRR610 pKa = 11.84GRR612 pKa = 11.84LNLGLPYY619 pKa = 9.97TAGGLNCSIPCDD631 pKa = 3.67YY632 pKa = 10.68RR633 pKa = 11.84VTDD636 pKa = 3.78GQRR639 pKa = 11.84RR640 pKa = 11.84LQRR643 pKa = 11.84YY644 pKa = 8.03CEE646 pKa = 3.47RR647 pKa = 11.84LYY649 pKa = 11.03RR650 pKa = 11.84NEE652 pKa = 3.31GRR654 pKa = 11.84TLTGVKK660 pKa = 9.92GRR662 pKa = 11.84SHH664 pKa = 6.71MLHH667 pKa = 5.32KK668 pKa = 10.27HH669 pKa = 4.54GRR671 pKa = 11.84SNNMRR676 pKa = 11.84LYY678 pKa = 10.5DD679 pKa = 3.51GTYY682 pKa = 10.29EE683 pKa = 4.35VILSSSEE690 pKa = 3.82GQEE693 pKa = 3.34QRR695 pKa = 11.84VPVEE699 pKa = 4.23LLDD702 pKa = 4.35APRR705 pKa = 11.84EE706 pKa = 4.12DD707 pKa = 3.72IILPVKK713 pKa = 10.48IPTEE717 pKa = 3.97THH719 pKa = 6.72LSPFGPILADD729 pKa = 3.82DD730 pKa = 3.77EE731 pKa = 4.91VEE733 pKa = 4.5NKK735 pKa = 9.91FSYY738 pKa = 10.27PSRR741 pKa = 11.84KK742 pKa = 10.31DD743 pKa = 2.89FDD745 pKa = 3.75QAKK748 pKa = 9.44KK749 pKa = 9.53FAPGGSPKK757 pKa = 10.38NFCLTKK763 pKa = 10.26KK764 pKa = 10.35VPYY767 pKa = 9.87DD768 pKa = 3.3KK769 pKa = 10.99RR770 pKa = 11.84FVTGKK775 pKa = 9.74VIKK778 pKa = 8.6VTPYY782 pKa = 10.67
Molecular weight: 89.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KI60|A0A1L3KI60_9VIRU Uncharacterized protein OS=Beihai narna-like virus 11 OX=1922438 PE=4 SV=1
MM1 pKa = 7.81SNITKK6 pKa = 10.45AKK8 pKa = 9.27FFQRR12 pKa = 11.84PSLKK16 pKa = 10.45GLPKK20 pKa = 9.87QEE22 pKa = 4.26KK23 pKa = 7.44EE24 pKa = 4.29RR25 pKa = 11.84RR26 pKa = 11.84WKK28 pKa = 8.91QHH30 pKa = 4.93LMSLGGEE37 pKa = 4.19TPRR40 pKa = 11.84NSRR43 pKa = 11.84RR44 pKa = 11.84PSRR47 pKa = 11.84RR48 pKa = 11.84NKK50 pKa = 9.07VAVGRR55 pKa = 11.84MKK57 pKa = 10.62TMPEE61 pKa = 3.9DD62 pKa = 3.7EE63 pKa = 4.58RR64 pKa = 11.84CGLEE68 pKa = 4.07YY69 pKa = 9.76ATSLINPFDD78 pKa = 4.11FSLNPCVPRR87 pKa = 11.84MPSVPSRR94 pKa = 11.84KK95 pKa = 9.24LCTYY99 pKa = 11.04ASGLGSTSGTTSYY112 pKa = 11.48GGVTAYY118 pKa = 7.47MTAANDD124 pKa = 3.86TTSVAVTEE132 pKa = 4.38TAFTGLSLPAWNSTGASSKK151 pKa = 8.0TQNSPYY157 pKa = 10.52SGADD161 pKa = 3.6FSANGAQVRR170 pKa = 11.84LVSAGLKK177 pKa = 10.05VRR179 pKa = 11.84FTGTKK184 pKa = 9.07LNQGGVCFPFLEE196 pKa = 5.01PDD198 pKa = 3.43LGDD201 pKa = 3.47VTGFTANDD209 pKa = 2.84IAAYY213 pKa = 9.6DD214 pKa = 3.49QYY216 pKa = 11.5FQGIEE221 pKa = 4.02FDD223 pKa = 4.17SDD225 pKa = 3.18WVSITFTPRR234 pKa = 11.84HH235 pKa = 5.73PADD238 pKa = 3.73FDD240 pKa = 3.55FAAADD245 pKa = 3.63KK246 pKa = 10.18PLTGQHH252 pKa = 6.37PCMGILIMSAAPNQPFEE269 pKa = 4.65YY270 pKa = 9.97VWTAHH275 pKa = 5.95WEE277 pKa = 4.44VIGRR281 pKa = 11.84NARR284 pKa = 11.84GKK286 pKa = 7.74TLSHH290 pKa = 7.23TYY292 pKa = 10.29PNTDD296 pKa = 3.77RR297 pKa = 11.84IISGASQFPPRR308 pKa = 11.84ALSAIQNAPRR318 pKa = 11.84PEE320 pKa = 3.93SLAAQVGRR328 pKa = 11.84NLMAHH333 pKa = 6.49GAGVVEE339 pKa = 4.18SLGRR343 pKa = 11.84GAMNMVGAAVADD355 pKa = 3.99TMVSGAAGLLSIGVV369 pKa = 3.65
MM1 pKa = 7.81SNITKK6 pKa = 10.45AKK8 pKa = 9.27FFQRR12 pKa = 11.84PSLKK16 pKa = 10.45GLPKK20 pKa = 9.87QEE22 pKa = 4.26KK23 pKa = 7.44EE24 pKa = 4.29RR25 pKa = 11.84RR26 pKa = 11.84WKK28 pKa = 8.91QHH30 pKa = 4.93LMSLGGEE37 pKa = 4.19TPRR40 pKa = 11.84NSRR43 pKa = 11.84RR44 pKa = 11.84PSRR47 pKa = 11.84RR48 pKa = 11.84NKK50 pKa = 9.07VAVGRR55 pKa = 11.84MKK57 pKa = 10.62TMPEE61 pKa = 3.9DD62 pKa = 3.7EE63 pKa = 4.58RR64 pKa = 11.84CGLEE68 pKa = 4.07YY69 pKa = 9.76ATSLINPFDD78 pKa = 4.11FSLNPCVPRR87 pKa = 11.84MPSVPSRR94 pKa = 11.84KK95 pKa = 9.24LCTYY99 pKa = 11.04ASGLGSTSGTTSYY112 pKa = 11.48GGVTAYY118 pKa = 7.47MTAANDD124 pKa = 3.86TTSVAVTEE132 pKa = 4.38TAFTGLSLPAWNSTGASSKK151 pKa = 8.0TQNSPYY157 pKa = 10.52SGADD161 pKa = 3.6FSANGAQVRR170 pKa = 11.84LVSAGLKK177 pKa = 10.05VRR179 pKa = 11.84FTGTKK184 pKa = 9.07LNQGGVCFPFLEE196 pKa = 5.01PDD198 pKa = 3.43LGDD201 pKa = 3.47VTGFTANDD209 pKa = 2.84IAAYY213 pKa = 9.6DD214 pKa = 3.49QYY216 pKa = 11.5FQGIEE221 pKa = 4.02FDD223 pKa = 4.17SDD225 pKa = 3.18WVSITFTPRR234 pKa = 11.84HH235 pKa = 5.73PADD238 pKa = 3.73FDD240 pKa = 3.55FAAADD245 pKa = 3.63KK246 pKa = 10.18PLTGQHH252 pKa = 6.37PCMGILIMSAAPNQPFEE269 pKa = 4.65YY270 pKa = 9.97VWTAHH275 pKa = 5.95WEE277 pKa = 4.44VIGRR281 pKa = 11.84NARR284 pKa = 11.84GKK286 pKa = 7.74TLSHH290 pKa = 7.23TYY292 pKa = 10.29PNTDD296 pKa = 3.77RR297 pKa = 11.84IISGASQFPPRR308 pKa = 11.84ALSAIQNAPRR318 pKa = 11.84PEE320 pKa = 3.93SLAAQVGRR328 pKa = 11.84NLMAHH333 pKa = 6.49GAGVVEE339 pKa = 4.18SLGRR343 pKa = 11.84GAMNMVGAAVADD355 pKa = 3.99TMVSGAAGLLSIGVV369 pKa = 3.65
Molecular weight: 39.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1151 |
369 |
782 |
575.5 |
64.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.69 ± 2.125 | 2.085 ± 0.374 |
5.039 ± 0.499 | 5.3 ± 0.909 |
3.91 ± 0.357 | 7.732 ± 0.897 |
2.259 ± 0.324 | 4.431 ± 0.603 |
5.213 ± 0.726 | 9.209 ± 1.246 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.172 ± 0.553 | 4.257 ± 0.179 |
6.082 ± 0.355 | 3.823 ± 0.153 |
7.124 ± 0.595 | 7.993 ± 0.486 |
5.387 ± 1.266 | 5.995 ± 0.017 |
1.564 ± 0.107 | 3.736 ± 0.664 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
