
Human papillomavirus 154
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 11
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
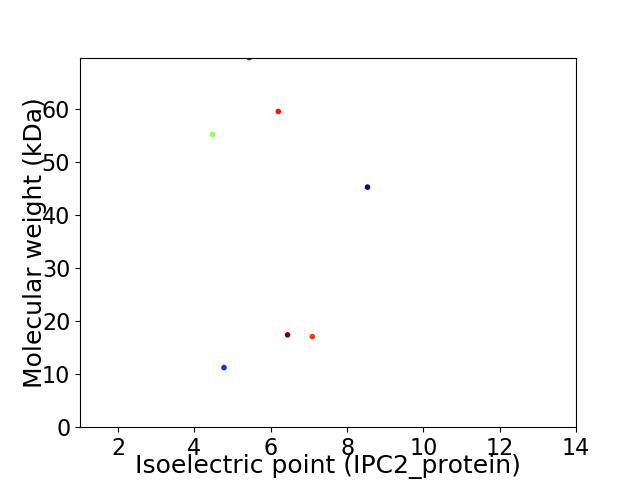
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9QBU3|R9QBU3_9PAPI Major capsid protein L1 OS=Human papillomavirus 154 OX=1195796 GN=L1 PE=3 SV=1
MM1 pKa = 7.28MGQTVTLKK9 pKa = 10.49DD10 pKa = 3.37IEE12 pKa = 4.49LGEE15 pKa = 4.19PLEE18 pKa = 4.36DD19 pKa = 3.46LVMPAHH25 pKa = 6.93LLCEE29 pKa = 4.28EE30 pKa = 4.73EE31 pKa = 4.81SLSSDD36 pKa = 3.28DD37 pKa = 3.91TPEE40 pKa = 4.15EE41 pKa = 4.26EE42 pKa = 4.82SLSPYY47 pKa = 10.42QVDD50 pKa = 4.78TICNSCNKK58 pKa = 10.24RR59 pKa = 11.84IRR61 pKa = 11.84LCVLATTGAIHH72 pKa = 7.33LLEE75 pKa = 3.86QLLFHH80 pKa = 7.18DD81 pKa = 5.32LSLVCPHH88 pKa = 7.28CSRR91 pKa = 11.84DD92 pKa = 3.32ILRR95 pKa = 11.84HH96 pKa = 4.98GRR98 pKa = 11.84THH100 pKa = 7.09
MM1 pKa = 7.28MGQTVTLKK9 pKa = 10.49DD10 pKa = 3.37IEE12 pKa = 4.49LGEE15 pKa = 4.19PLEE18 pKa = 4.36DD19 pKa = 3.46LVMPAHH25 pKa = 6.93LLCEE29 pKa = 4.28EE30 pKa = 4.73EE31 pKa = 4.81SLSSDD36 pKa = 3.28DD37 pKa = 3.91TPEE40 pKa = 4.15EE41 pKa = 4.26EE42 pKa = 4.82SLSPYY47 pKa = 10.42QVDD50 pKa = 4.78TICNSCNKK58 pKa = 10.24RR59 pKa = 11.84IRR61 pKa = 11.84LCVLATTGAIHH72 pKa = 7.33LLEE75 pKa = 3.86QLLFHH80 pKa = 7.18DD81 pKa = 5.32LSLVCPHH88 pKa = 7.28CSRR91 pKa = 11.84DD92 pKa = 3.32ILRR95 pKa = 11.84HH96 pKa = 4.98GRR98 pKa = 11.84THH100 pKa = 7.09
Molecular weight: 11.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9QBU2|R9QBU2_9PAPI Protein E7 OS=Human papillomavirus 154 OX=1195796 GN=E7 PE=3 SV=1
MM1 pKa = 6.96NQNDD5 pKa = 3.41LRR7 pKa = 11.84EE8 pKa = 4.28RR9 pKa = 11.84LDD11 pKa = 3.72ALQEE15 pKa = 4.03QLMTLYY21 pKa = 10.65EE22 pKa = 4.67LGPTDD27 pKa = 4.5LSSQIKK33 pKa = 9.96HH34 pKa = 4.91YY35 pKa = 10.56QLLRR39 pKa = 11.84KK40 pKa = 9.61EE41 pKa = 4.53SVLEE45 pKa = 3.79YY46 pKa = 9.75YY47 pKa = 10.17ARR49 pKa = 11.84KK50 pKa = 9.55EE51 pKa = 4.12GYY53 pKa = 9.04EE54 pKa = 3.98VLGLHH59 pKa = 6.62HH60 pKa = 7.21LPVLKK65 pKa = 10.76VSEE68 pKa = 4.69HH69 pKa = 5.67NAKK72 pKa = 9.86QAIKK76 pKa = 9.71MILHH80 pKa = 6.75LEE82 pKa = 4.13SLAKK86 pKa = 9.83SAYY89 pKa = 9.31KK90 pKa = 10.62HH91 pKa = 5.27EE92 pKa = 4.35TWTLNDD98 pKa = 3.32TSADD102 pKa = 3.31RR103 pKa = 11.84FMSPPRR109 pKa = 11.84NCFKK113 pKa = 10.69KK114 pKa = 10.55DD115 pKa = 3.39SFEE118 pKa = 4.03VTVWFDD124 pKa = 3.93HH125 pKa = 6.16NPQNAFPYY133 pKa = 10.45INWKK137 pKa = 8.35WIYY140 pKa = 10.29YY141 pKa = 9.44QDD143 pKa = 5.88NNDD146 pKa = 3.09QWHH149 pKa = 6.23KK150 pKa = 10.53VPGRR154 pKa = 11.84TDD156 pKa = 3.08HH157 pKa = 6.55NGLYY161 pKa = 10.37FIEE164 pKa = 5.05HH165 pKa = 7.62DD166 pKa = 3.6NTVTYY171 pKa = 10.41FLLFAKK177 pKa = 10.44DD178 pKa = 3.2AEE180 pKa = 4.63RR181 pKa = 11.84YY182 pKa = 9.36GKK184 pKa = 8.26TKK186 pKa = 10.04EE187 pKa = 3.67WTVNIGNEE195 pKa = 4.23QILPPSVTSSSRR207 pKa = 11.84RR208 pKa = 11.84SLSDD212 pKa = 3.12SPEE215 pKa = 4.09VNRR218 pKa = 11.84GASTSTYY225 pKa = 11.16AEE227 pKa = 3.9AEE229 pKa = 4.1EE230 pKa = 4.13EE231 pKa = 4.01NRR233 pKa = 11.84RR234 pKa = 11.84GSLQPQTTSPSTTTYY249 pKa = 9.75SPSRR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84GQGEE262 pKa = 4.14STTNKK267 pKa = 8.67RR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84AEE273 pKa = 3.74RR274 pKa = 11.84DD275 pKa = 2.98GGADD279 pKa = 2.99TVSPDD284 pKa = 3.43EE285 pKa = 4.51VGQSHH290 pKa = 5.71QLVRR294 pKa = 11.84STGLTRR300 pKa = 11.84TEE302 pKa = 3.97RR303 pKa = 11.84LKK305 pKa = 11.21EE306 pKa = 3.89EE307 pKa = 4.7AKK309 pKa = 10.77DD310 pKa = 3.69PPIVSISGPANKK322 pKa = 9.8LKK324 pKa = 10.19CWRR327 pKa = 11.84YY328 pKa = 9.06RR329 pKa = 11.84CGLKK333 pKa = 9.38TSKK336 pKa = 10.57NYY338 pKa = 10.34TYY340 pKa = 10.06MSSVFKK346 pKa = 10.46WITGDD351 pKa = 3.21VGLGDD356 pKa = 3.68GRR358 pKa = 11.84MLVAFANTQQRR369 pKa = 11.84NQFINALTLPKK380 pKa = 10.75DD381 pKa = 3.3MSYY384 pKa = 11.41CLGNLEE390 pKa = 4.54KK391 pKa = 10.97LL392 pKa = 3.77
MM1 pKa = 6.96NQNDD5 pKa = 3.41LRR7 pKa = 11.84EE8 pKa = 4.28RR9 pKa = 11.84LDD11 pKa = 3.72ALQEE15 pKa = 4.03QLMTLYY21 pKa = 10.65EE22 pKa = 4.67LGPTDD27 pKa = 4.5LSSQIKK33 pKa = 9.96HH34 pKa = 4.91YY35 pKa = 10.56QLLRR39 pKa = 11.84KK40 pKa = 9.61EE41 pKa = 4.53SVLEE45 pKa = 3.79YY46 pKa = 9.75YY47 pKa = 10.17ARR49 pKa = 11.84KK50 pKa = 9.55EE51 pKa = 4.12GYY53 pKa = 9.04EE54 pKa = 3.98VLGLHH59 pKa = 6.62HH60 pKa = 7.21LPVLKK65 pKa = 10.76VSEE68 pKa = 4.69HH69 pKa = 5.67NAKK72 pKa = 9.86QAIKK76 pKa = 9.71MILHH80 pKa = 6.75LEE82 pKa = 4.13SLAKK86 pKa = 9.83SAYY89 pKa = 9.31KK90 pKa = 10.62HH91 pKa = 5.27EE92 pKa = 4.35TWTLNDD98 pKa = 3.32TSADD102 pKa = 3.31RR103 pKa = 11.84FMSPPRR109 pKa = 11.84NCFKK113 pKa = 10.69KK114 pKa = 10.55DD115 pKa = 3.39SFEE118 pKa = 4.03VTVWFDD124 pKa = 3.93HH125 pKa = 6.16NPQNAFPYY133 pKa = 10.45INWKK137 pKa = 8.35WIYY140 pKa = 10.29YY141 pKa = 9.44QDD143 pKa = 5.88NNDD146 pKa = 3.09QWHH149 pKa = 6.23KK150 pKa = 10.53VPGRR154 pKa = 11.84TDD156 pKa = 3.08HH157 pKa = 6.55NGLYY161 pKa = 10.37FIEE164 pKa = 5.05HH165 pKa = 7.62DD166 pKa = 3.6NTVTYY171 pKa = 10.41FLLFAKK177 pKa = 10.44DD178 pKa = 3.2AEE180 pKa = 4.63RR181 pKa = 11.84YY182 pKa = 9.36GKK184 pKa = 8.26TKK186 pKa = 10.04EE187 pKa = 3.67WTVNIGNEE195 pKa = 4.23QILPPSVTSSSRR207 pKa = 11.84RR208 pKa = 11.84SLSDD212 pKa = 3.12SPEE215 pKa = 4.09VNRR218 pKa = 11.84GASTSTYY225 pKa = 11.16AEE227 pKa = 3.9AEE229 pKa = 4.1EE230 pKa = 4.13EE231 pKa = 4.01NRR233 pKa = 11.84RR234 pKa = 11.84GSLQPQTTSPSTTTYY249 pKa = 9.75SPSRR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84GQGEE262 pKa = 4.14STTNKK267 pKa = 8.67RR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84AEE273 pKa = 3.74RR274 pKa = 11.84DD275 pKa = 2.98GGADD279 pKa = 2.99TVSPDD284 pKa = 3.43EE285 pKa = 4.51VGQSHH290 pKa = 5.71QLVRR294 pKa = 11.84STGLTRR300 pKa = 11.84TEE302 pKa = 3.97RR303 pKa = 11.84LKK305 pKa = 11.21EE306 pKa = 3.89EE307 pKa = 4.7AKK309 pKa = 10.77DD310 pKa = 3.69PPIVSISGPANKK322 pKa = 9.8LKK324 pKa = 10.19CWRR327 pKa = 11.84YY328 pKa = 9.06RR329 pKa = 11.84CGLKK333 pKa = 9.38TSKK336 pKa = 10.57NYY338 pKa = 10.34TYY340 pKa = 10.06MSSVFKK346 pKa = 10.46WITGDD351 pKa = 3.21VGLGDD356 pKa = 3.68GRR358 pKa = 11.84MLVAFANTQQRR369 pKa = 11.84NQFINALTLPKK380 pKa = 10.75DD381 pKa = 3.3MSYY384 pKa = 11.41CLGNLEE390 pKa = 4.54KK391 pKa = 10.97LL392 pKa = 3.77
Molecular weight: 45.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2422 |
100 |
606 |
346.0 |
39.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.202 ± 0.443 | 2.271 ± 0.709 |
6.4 ± 0.302 | 6.235 ± 0.586 |
4.707 ± 0.61 | 5.326 ± 0.745 |
2.147 ± 0.395 | 5.822 ± 0.921 |
5.285 ± 0.701 | 9.29 ± 1.154 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.982 ± 0.283 | 5.409 ± 0.63 |
6.482 ± 0.826 | 4.583 ± 0.372 |
5.615 ± 0.685 | 7.019 ± 0.707 |
6.358 ± 0.469 | 5.202 ± 0.61 |
1.239 ± 0.237 | 3.427 ± 0.447 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
