
Hypericum japonicum associated circular DNA virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus hypas1; Hypericum associated gemycircularvirus 1
Average proteome isoelectric point is 7.78
Get precalculated fractions of proteins
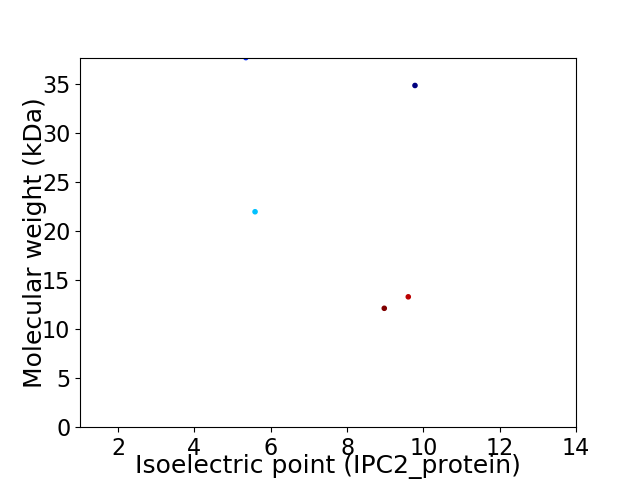
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5S0Z8|S5S0Z8_9VIRU Putative V2 protein OS=Hypericum japonicum associated circular DNA virus OX=1367218 PE=4 SV=1
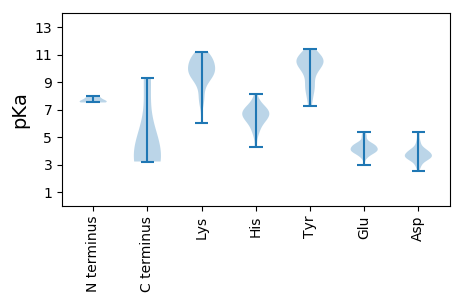
MM1 pKa = 7.51PQLEE5 pKa = 4.38WNFRR9 pKa = 11.84YY10 pKa = 10.49VLVTYY15 pKa = 7.86AQCGDD20 pKa = 3.91LDD22 pKa = 3.45PWRR25 pKa = 11.84VVEE28 pKa = 4.5RR29 pKa = 11.84FSSLGAEE36 pKa = 4.2CIVGRR41 pKa = 11.84EE42 pKa = 3.95HH43 pKa = 8.11HH44 pKa = 6.88EE45 pKa = 4.43DD46 pKa = 3.53GGLHH50 pKa = 5.69LHH52 pKa = 6.24VFADD56 pKa = 4.7FGRR59 pKa = 11.84KK60 pKa = 8.18FRR62 pKa = 11.84SRR64 pKa = 11.84KK65 pKa = 9.81ADD67 pKa = 3.2ILDD70 pKa = 3.23VDD72 pKa = 3.98GRR74 pKa = 11.84HH75 pKa = 6.5PNLAPIKK82 pKa = 8.35RR83 pKa = 11.84TPAKK87 pKa = 10.37AYY89 pKa = 10.34DD90 pKa = 3.71YY91 pKa = 10.64AIKK94 pKa = 10.92DD95 pKa = 3.52GDD97 pKa = 4.01VVAGGLEE104 pKa = 3.95RR105 pKa = 11.84PVEE108 pKa = 4.29GGVGNGTSADD118 pKa = 2.91KK119 pKa = 10.31WSRR122 pKa = 11.84ITQAEE127 pKa = 4.09DD128 pKa = 3.68RR129 pKa = 11.84DD130 pKa = 4.22EE131 pKa = 4.72FWALVHH137 pKa = 6.48EE138 pKa = 5.38LDD140 pKa = 4.56PKK142 pKa = 10.9AAACSFNALSKK153 pKa = 9.33YY154 pKa = 10.43ADD156 pKa = 2.87WRR158 pKa = 11.84FAEE161 pKa = 4.39KK162 pKa = 10.48PPVYY166 pKa = 9.95EE167 pKa = 3.52HH168 pKa = 7.52DD169 pKa = 3.54GRR171 pKa = 11.84IEE173 pKa = 4.01FVPGDD178 pKa = 3.54ADD180 pKa = 5.19GRR182 pKa = 11.84DD183 pKa = 3.5DD184 pKa = 3.59WVSQSGIGLDD194 pKa = 3.7EE195 pKa = 4.64PFLGGRR201 pKa = 11.84VKK203 pKa = 10.9SLVLFGGTRR212 pKa = 11.84TGKK215 pKa = 5.99TTWARR220 pKa = 11.84SLGKK224 pKa = 9.48HH225 pKa = 6.3LYY227 pKa = 10.6CIGLVSGAEE236 pKa = 4.08CSKK239 pKa = 11.2GADD242 pKa = 2.93AEE244 pKa = 4.49YY245 pKa = 10.97AVFDD249 pKa = 5.21DD250 pKa = 3.48IRR252 pKa = 11.84GGFGFFHH259 pKa = 7.53GYY261 pKa = 9.92KK262 pKa = 9.15EE263 pKa = 3.99WLGAQPHH270 pKa = 5.31VSIKK274 pKa = 9.68QLYY277 pKa = 8.59RR278 pKa = 11.84EE279 pKa = 4.34PYY281 pKa = 7.28YY282 pKa = 10.19MKK284 pKa = 9.88WGKK287 pKa = 9.51PSIWICNTDD296 pKa = 3.34PRR298 pKa = 11.84LDD300 pKa = 3.53AYY302 pKa = 10.49GPNATPDD309 pKa = 3.75WEE311 pKa = 4.2WMEE314 pKa = 4.4GNCHH318 pKa = 6.54FIEE321 pKa = 4.61VKK323 pKa = 10.61DD324 pKa = 3.86SLISSISHH332 pKa = 7.12ASTEE336 pKa = 4.06
MM1 pKa = 7.51PQLEE5 pKa = 4.38WNFRR9 pKa = 11.84YY10 pKa = 10.49VLVTYY15 pKa = 7.86AQCGDD20 pKa = 3.91LDD22 pKa = 3.45PWRR25 pKa = 11.84VVEE28 pKa = 4.5RR29 pKa = 11.84FSSLGAEE36 pKa = 4.2CIVGRR41 pKa = 11.84EE42 pKa = 3.95HH43 pKa = 8.11HH44 pKa = 6.88EE45 pKa = 4.43DD46 pKa = 3.53GGLHH50 pKa = 5.69LHH52 pKa = 6.24VFADD56 pKa = 4.7FGRR59 pKa = 11.84KK60 pKa = 8.18FRR62 pKa = 11.84SRR64 pKa = 11.84KK65 pKa = 9.81ADD67 pKa = 3.2ILDD70 pKa = 3.23VDD72 pKa = 3.98GRR74 pKa = 11.84HH75 pKa = 6.5PNLAPIKK82 pKa = 8.35RR83 pKa = 11.84TPAKK87 pKa = 10.37AYY89 pKa = 10.34DD90 pKa = 3.71YY91 pKa = 10.64AIKK94 pKa = 10.92DD95 pKa = 3.52GDD97 pKa = 4.01VVAGGLEE104 pKa = 3.95RR105 pKa = 11.84PVEE108 pKa = 4.29GGVGNGTSADD118 pKa = 2.91KK119 pKa = 10.31WSRR122 pKa = 11.84ITQAEE127 pKa = 4.09DD128 pKa = 3.68RR129 pKa = 11.84DD130 pKa = 4.22EE131 pKa = 4.72FWALVHH137 pKa = 6.48EE138 pKa = 5.38LDD140 pKa = 4.56PKK142 pKa = 10.9AAACSFNALSKK153 pKa = 9.33YY154 pKa = 10.43ADD156 pKa = 2.87WRR158 pKa = 11.84FAEE161 pKa = 4.39KK162 pKa = 10.48PPVYY166 pKa = 9.95EE167 pKa = 3.52HH168 pKa = 7.52DD169 pKa = 3.54GRR171 pKa = 11.84IEE173 pKa = 4.01FVPGDD178 pKa = 3.54ADD180 pKa = 5.19GRR182 pKa = 11.84DD183 pKa = 3.5DD184 pKa = 3.59WVSQSGIGLDD194 pKa = 3.7EE195 pKa = 4.64PFLGGRR201 pKa = 11.84VKK203 pKa = 10.9SLVLFGGTRR212 pKa = 11.84TGKK215 pKa = 5.99TTWARR220 pKa = 11.84SLGKK224 pKa = 9.48HH225 pKa = 6.3LYY227 pKa = 10.6CIGLVSGAEE236 pKa = 4.08CSKK239 pKa = 11.2GADD242 pKa = 2.93AEE244 pKa = 4.49YY245 pKa = 10.97AVFDD249 pKa = 5.21DD250 pKa = 3.48IRR252 pKa = 11.84GGFGFFHH259 pKa = 7.53GYY261 pKa = 9.92KK262 pKa = 9.15EE263 pKa = 3.99WLGAQPHH270 pKa = 5.31VSIKK274 pKa = 9.68QLYY277 pKa = 8.59RR278 pKa = 11.84EE279 pKa = 4.34PYY281 pKa = 7.28YY282 pKa = 10.19MKK284 pKa = 9.88WGKK287 pKa = 9.51PSIWICNTDD296 pKa = 3.34PRR298 pKa = 11.84LDD300 pKa = 3.53AYY302 pKa = 10.49GPNATPDD309 pKa = 3.75WEE311 pKa = 4.2WMEE314 pKa = 4.4GNCHH318 pKa = 6.54FIEE321 pKa = 4.61VKK323 pKa = 10.61DD324 pKa = 3.86SLISSISHH332 pKa = 7.12ASTEE336 pKa = 4.06
Molecular weight: 37.64 kDa
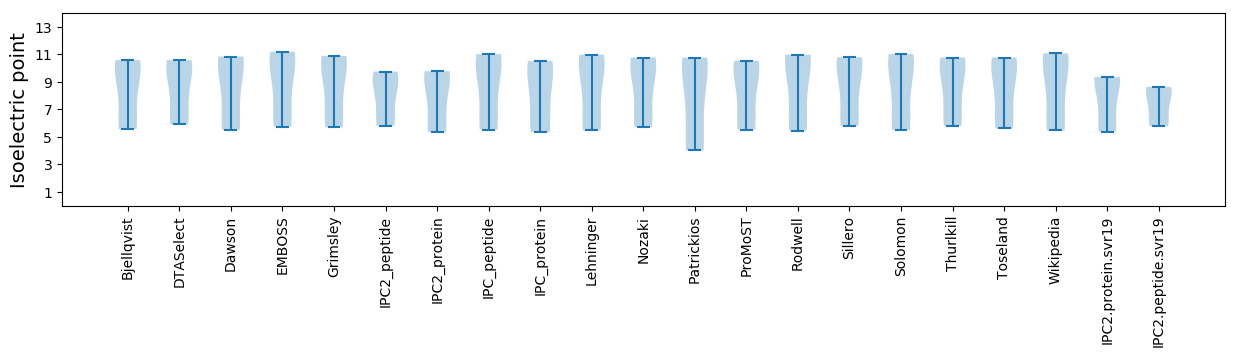
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5S0Z8|S5S0Z8_9VIRU Putative V2 protein OS=Hypericum japonicum associated circular DNA virus OX=1367218 PE=4 SV=1
MM1 pKa = 7.79APMLVILLHH10 pKa = 6.93CYY12 pKa = 10.31LKK14 pKa = 10.86LLCTGMRR21 pKa = 11.84NRR23 pKa = 11.84RR24 pKa = 11.84DD25 pKa = 3.34KK26 pKa = 11.09RR27 pKa = 11.84VLDD30 pKa = 4.1FNEE33 pKa = 3.99MTIPLHH39 pKa = 5.85PFPIRR44 pKa = 11.84RR45 pKa = 11.84SVRR48 pKa = 11.84AIGIQTRR55 pKa = 11.84VGVANPNRR63 pKa = 11.84RR64 pKa = 11.84LTPLHH69 pKa = 6.27VVGLPVKK76 pKa = 10.64LLDD79 pKa = 3.92GYY81 pKa = 10.24MRR83 pKa = 11.84LSTEE87 pKa = 3.91PLLVSVKK94 pKa = 9.61EE95 pKa = 4.39SKK97 pKa = 7.47TTSDD101 pKa = 3.48VVEE104 pKa = 5.04DD105 pKa = 4.11GVLGVGSLRR114 pKa = 11.84TFRR117 pKa = 11.84TT118 pKa = 3.52
MM1 pKa = 7.79APMLVILLHH10 pKa = 6.93CYY12 pKa = 10.31LKK14 pKa = 10.86LLCTGMRR21 pKa = 11.84NRR23 pKa = 11.84RR24 pKa = 11.84DD25 pKa = 3.34KK26 pKa = 11.09RR27 pKa = 11.84VLDD30 pKa = 4.1FNEE33 pKa = 3.99MTIPLHH39 pKa = 5.85PFPIRR44 pKa = 11.84RR45 pKa = 11.84SVRR48 pKa = 11.84AIGIQTRR55 pKa = 11.84VGVANPNRR63 pKa = 11.84RR64 pKa = 11.84LTPLHH69 pKa = 6.27VVGLPVKK76 pKa = 10.64LLDD79 pKa = 3.92GYY81 pKa = 10.24MRR83 pKa = 11.84LSTEE87 pKa = 3.91PLLVSVKK94 pKa = 9.61EE95 pKa = 4.39SKK97 pKa = 7.47TTSDD101 pKa = 3.48VVEE104 pKa = 5.04DD105 pKa = 4.11GVLGVGSLRR114 pKa = 11.84TFRR117 pKa = 11.84TT118 pKa = 3.52
Molecular weight: 13.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1052 |
106 |
336 |
210.4 |
23.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.703 ± 1.19 | 1.521 ± 0.385 |
5.513 ± 1.03 | 5.323 ± 1.34 |
3.232 ± 0.589 | 7.319 ± 1.383 |
2.567 ± 0.691 | 4.373 ± 0.281 |
4.183 ± 0.793 | 9.125 ± 1.307 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.327 ± 1.041 | 4.183 ± 1.334 |
4.848 ± 0.52 | 2.376 ± 0.469 |
8.84 ± 0.926 | 8.365 ± 2.12 |
6.939 ± 1.667 | 5.989 ± 1.204 |
2.852 ± 0.502 | 3.422 ± 0.872 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
