
Pelagibaculum spongiae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Oceanospirillales incertae sedis; Pelagibaculum
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
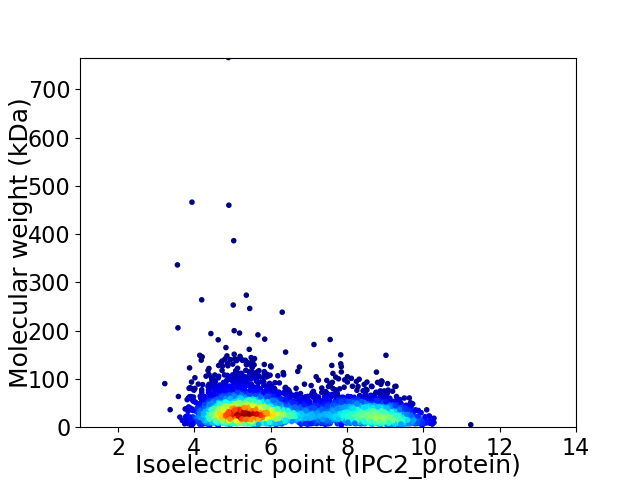
Virtual 2D-PAGE plot for 4189 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V1GZJ2|A0A2V1GZJ2_9GAMM DUF3307 domain-containing protein OS=Pelagibaculum spongiae OX=2080658 GN=DC094_03845 PE=4 SV=1
MM1 pKa = 7.6KK2 pKa = 10.23KK3 pKa = 10.75GSFLKK8 pKa = 10.48RR9 pKa = 11.84SAVVAFSVLLMAGCGGSGVTDD30 pKa = 3.56TTSGGGTDD38 pKa = 3.62GGTDD42 pKa = 3.5GGTVTPPTTQPFATTCVGTGSSATLGFVNFDD73 pKa = 3.54GASSLEE79 pKa = 4.03LAQKK83 pKa = 10.7GSQSDD88 pKa = 3.64NRR90 pKa = 11.84PQSSTVVFSACSEE103 pKa = 4.39DD104 pKa = 3.58GTPVAGLTVSFSLSTSSGGIAISQASATTDD134 pKa = 3.21ANGQASTTITSGTTATAFILAASVQQSSITEE165 pKa = 3.99TKK167 pKa = 10.33NQVFTIGGSVPDD179 pKa = 4.38ANSFSLSQAATNVDD193 pKa = 3.49GYY195 pKa = 10.96EE196 pKa = 4.02FDD198 pKa = 4.01GATAGINIRR207 pKa = 11.84LADD210 pKa = 3.75HH211 pKa = 6.54VNNPVQDD218 pKa = 3.83GTPVSFITNGGAIQSSCPTALAGGTASCSVTWEE251 pKa = 4.17SQDD254 pKa = 3.61PQPCYY259 pKa = 10.55YY260 pKa = 10.53EE261 pKa = 4.22QVTLSAAQTGLTGIDD276 pKa = 3.05INEE279 pKa = 4.0AVAVCGRR286 pKa = 11.84AAVLAWTEE294 pKa = 4.29GEE296 pKa = 4.27EE297 pKa = 4.92SFNDD301 pKa = 3.46PNQNGVYY308 pKa = 10.21DD309 pKa = 3.9AGEE312 pKa = 4.06TVIGVNNQPGGDD324 pKa = 3.62LAEE327 pKa = 5.4PYY329 pKa = 10.89LDD331 pKa = 3.69VNFNGQYY338 pKa = 10.64DD339 pKa = 3.49IAEE342 pKa = 4.38AFVDD346 pKa = 3.91ANGNSAFDD354 pKa = 3.86PSEE357 pKa = 4.06TYY359 pKa = 10.21TDD361 pKa = 3.66SNNNGVYY368 pKa = 10.48DD369 pKa = 3.83SAEE372 pKa = 4.47PIVDD376 pKa = 3.47RR377 pKa = 11.84DD378 pKa = 3.44GDD380 pKa = 3.94GTRR383 pKa = 11.84TAGDD387 pKa = 3.17AKK389 pKa = 10.32YY390 pKa = 9.07TGLRR394 pKa = 11.84CEE396 pKa = 4.39EE397 pKa = 4.36TNASCTRR404 pKa = 11.84GTLFLYY410 pKa = 9.76DD411 pKa = 3.23QRR413 pKa = 11.84TVVMSGRR420 pKa = 11.84TINARR425 pKa = 11.84ILPASARR432 pKa = 11.84PIIQAAVARR441 pKa = 11.84SSYY444 pKa = 10.54AQTIAILDD452 pKa = 3.88KK453 pKa = 10.89LVAEE457 pKa = 4.65ATPYY461 pKa = 10.35TYY463 pKa = 11.15GDD465 pKa = 3.99EE466 pKa = 4.26LTLLVQDD473 pKa = 4.14QNGNPLAAGTTIAIAAEE490 pKa = 4.3DD491 pKa = 3.68VDD493 pKa = 5.24MIPAAPSATLGQTATLEE510 pKa = 4.34NGQSWLNFTLVQPDD524 pKa = 4.15GGSGTPPEE532 pKa = 4.69EE533 pKa = 4.06GTLTVTITVSRR544 pKa = 11.84ADD546 pKa = 3.62SGVSTLAITYY556 pKa = 9.34VVEE559 pKa = 4.36DD560 pKa = 3.58
MM1 pKa = 7.6KK2 pKa = 10.23KK3 pKa = 10.75GSFLKK8 pKa = 10.48RR9 pKa = 11.84SAVVAFSVLLMAGCGGSGVTDD30 pKa = 3.56TTSGGGTDD38 pKa = 3.62GGTDD42 pKa = 3.5GGTVTPPTTQPFATTCVGTGSSATLGFVNFDD73 pKa = 3.54GASSLEE79 pKa = 4.03LAQKK83 pKa = 10.7GSQSDD88 pKa = 3.64NRR90 pKa = 11.84PQSSTVVFSACSEE103 pKa = 4.39DD104 pKa = 3.58GTPVAGLTVSFSLSTSSGGIAISQASATTDD134 pKa = 3.21ANGQASTTITSGTTATAFILAASVQQSSITEE165 pKa = 3.99TKK167 pKa = 10.33NQVFTIGGSVPDD179 pKa = 4.38ANSFSLSQAATNVDD193 pKa = 3.49GYY195 pKa = 10.96EE196 pKa = 4.02FDD198 pKa = 4.01GATAGINIRR207 pKa = 11.84LADD210 pKa = 3.75HH211 pKa = 6.54VNNPVQDD218 pKa = 3.83GTPVSFITNGGAIQSSCPTALAGGTASCSVTWEE251 pKa = 4.17SQDD254 pKa = 3.61PQPCYY259 pKa = 10.55YY260 pKa = 10.53EE261 pKa = 4.22QVTLSAAQTGLTGIDD276 pKa = 3.05INEE279 pKa = 4.0AVAVCGRR286 pKa = 11.84AAVLAWTEE294 pKa = 4.29GEE296 pKa = 4.27EE297 pKa = 4.92SFNDD301 pKa = 3.46PNQNGVYY308 pKa = 10.21DD309 pKa = 3.9AGEE312 pKa = 4.06TVIGVNNQPGGDD324 pKa = 3.62LAEE327 pKa = 5.4PYY329 pKa = 10.89LDD331 pKa = 3.69VNFNGQYY338 pKa = 10.64DD339 pKa = 3.49IAEE342 pKa = 4.38AFVDD346 pKa = 3.91ANGNSAFDD354 pKa = 3.86PSEE357 pKa = 4.06TYY359 pKa = 10.21TDD361 pKa = 3.66SNNNGVYY368 pKa = 10.48DD369 pKa = 3.83SAEE372 pKa = 4.47PIVDD376 pKa = 3.47RR377 pKa = 11.84DD378 pKa = 3.44GDD380 pKa = 3.94GTRR383 pKa = 11.84TAGDD387 pKa = 3.17AKK389 pKa = 10.32YY390 pKa = 9.07TGLRR394 pKa = 11.84CEE396 pKa = 4.39EE397 pKa = 4.36TNASCTRR404 pKa = 11.84GTLFLYY410 pKa = 9.76DD411 pKa = 3.23QRR413 pKa = 11.84TVVMSGRR420 pKa = 11.84TINARR425 pKa = 11.84ILPASARR432 pKa = 11.84PIIQAAVARR441 pKa = 11.84SSYY444 pKa = 10.54AQTIAILDD452 pKa = 3.88KK453 pKa = 10.89LVAEE457 pKa = 4.65ATPYY461 pKa = 10.35TYY463 pKa = 11.15GDD465 pKa = 3.99EE466 pKa = 4.26LTLLVQDD473 pKa = 4.14QNGNPLAAGTTIAIAAEE490 pKa = 4.3DD491 pKa = 3.68VDD493 pKa = 5.24MIPAAPSATLGQTATLEE510 pKa = 4.34NGQSWLNFTLVQPDD524 pKa = 4.15GGSGTPPEE532 pKa = 4.69EE533 pKa = 4.06GTLTVTITVSRR544 pKa = 11.84ADD546 pKa = 3.62SGVSTLAITYY556 pKa = 9.34VVEE559 pKa = 4.36DD560 pKa = 3.58
Molecular weight: 57.11 kDa
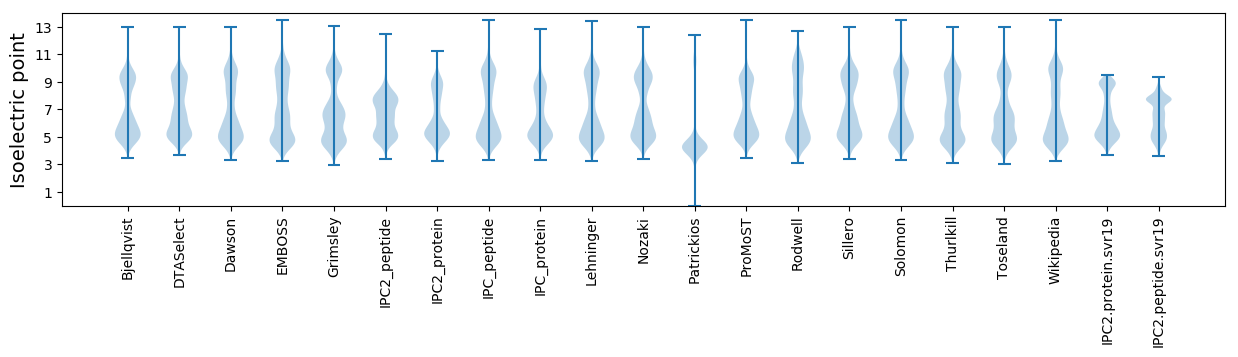
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V1GS63|A0A2V1GS63_9GAMM Endonuclease OS=Pelagibaculum spongiae OX=2080658 GN=DC094_17660 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38GGRR28 pKa = 11.84LVLKK32 pKa = 10.27RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.42GRR39 pKa = 11.84ARR41 pKa = 11.84LAAA44 pKa = 4.44
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38GGRR28 pKa = 11.84LVLKK32 pKa = 10.27RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.42GRR39 pKa = 11.84ARR41 pKa = 11.84LAAA44 pKa = 4.44
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1371512 |
26 |
6845 |
327.4 |
36.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.604 ± 0.043 | 1.165 ± 0.015 |
5.605 ± 0.039 | 5.842 ± 0.043 |
4.067 ± 0.023 | 6.694 ± 0.035 |
2.099 ± 0.018 | 6.307 ± 0.028 |
5.387 ± 0.04 | 10.802 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.461 ± 0.023 | 4.299 ± 0.037 |
4.141 ± 0.023 | 5.51 ± 0.057 |
4.773 ± 0.034 | 7.074 ± 0.034 |
4.821 ± 0.028 | 6.206 ± 0.035 |
1.34 ± 0.015 | 2.804 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
