
Angomonas deanei
Taxonomy: cellular organisms; Eukaryota; Discoba; Euglenozoa; Kinetoplastea; Metakinetoplastina; Trypanosomatida; Trypanosomatidae; Strigomonadinae; Angomonas
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
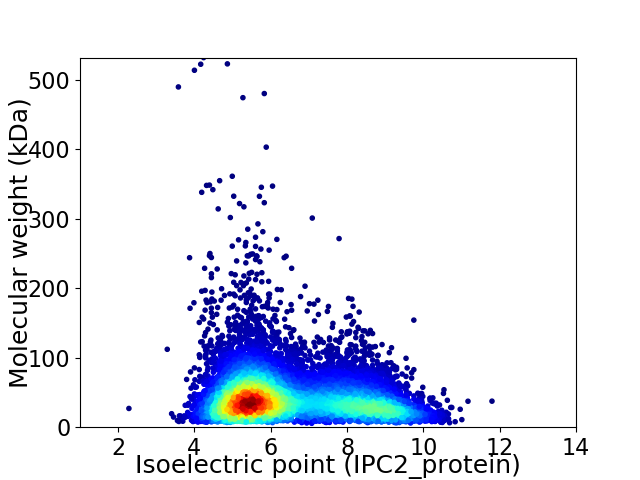
Virtual 2D-PAGE plot for 10115 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7G2CJY9|A0A7G2CJY9_9TRYP Uncharacterized protein OS=Angomonas deanei OX=59799 GN=ADEAN_000769300 PE=4 SV=1
MM1 pKa = 7.08FKK3 pKa = 10.78VMRR6 pKa = 11.84TLLLSLLLLATTLHH20 pKa = 6.6ASSSDD25 pKa = 3.36SGSNSTDD32 pKa = 2.96SGSNSTDD39 pKa = 2.96SGSNSTDD46 pKa = 2.96SGSNSTDD53 pKa = 2.98SSDD56 pKa = 4.92SYY58 pKa = 11.45LDD60 pKa = 3.51HH61 pKa = 7.16NKK63 pKa = 9.03TDD65 pKa = 3.67SGSGSHH71 pKa = 6.84HH72 pKa = 6.84SDD74 pKa = 2.97SGNSPDD80 pKa = 4.13SNHH83 pKa = 6.41SGSLPTDD90 pKa = 3.55SSADD94 pKa = 3.54DD95 pKa = 3.83SALSDD100 pKa = 4.48SDD102 pKa = 5.76DD103 pKa = 3.72GVCNVTHH110 pKa = 6.87CKK112 pKa = 10.04EE113 pKa = 3.88CAKK116 pKa = 10.74RR117 pKa = 11.84NVCLQCEE124 pKa = 4.28DD125 pKa = 4.65HH126 pKa = 6.9YY127 pKa = 11.63VLPPSSSTRR136 pKa = 11.84CVPVMDD142 pKa = 4.31GCLVYY147 pKa = 9.96TVNANVCQQCKK158 pKa = 9.86EE159 pKa = 4.2GYY161 pKa = 7.53TLEE164 pKa = 4.34TGGTCTGTKK173 pKa = 9.92KK174 pKa = 10.74KK175 pKa = 10.57EE176 pKa = 3.91GTQWWVWLIVALAIVVVIVIIIIIICCCCCKK207 pKa = 9.98KK208 pKa = 10.86DD209 pKa = 3.36KK210 pKa = 7.8TTPTRR215 pKa = 11.84GTSTTTSRR223 pKa = 11.84STSTNSSFSSKK234 pKa = 9.16TATGEE239 pKa = 4.05DD240 pKa = 4.36DD241 pKa = 4.55SDD243 pKa = 5.37SSDD246 pKa = 5.77DD247 pKa = 6.14DD248 pKa = 6.46DD249 pKa = 7.69DD250 pKa = 7.65DD251 pKa = 7.64DD252 pKa = 7.67DD253 pKa = 7.6DD254 pKa = 7.56DD255 pKa = 7.19DD256 pKa = 7.37DD257 pKa = 5.61EE258 pKa = 7.83DD259 pKa = 6.39SDD261 pKa = 6.34DD262 pKa = 4.81DD263 pKa = 6.15DD264 pKa = 5.15EE265 pKa = 7.02DD266 pKa = 4.48SSEE269 pKa = 4.27DD270 pKa = 3.47SSEE273 pKa = 3.98EE274 pKa = 3.84SSGDD278 pKa = 3.65EE279 pKa = 4.5EE280 pKa = 4.72EE281 pKa = 5.07DD282 pKa = 4.3SSDD285 pKa = 3.48EE286 pKa = 4.1SDD288 pKa = 3.91AEE290 pKa = 4.55GSAGTAEE297 pKa = 4.13ATVV300 pKa = 3.27
MM1 pKa = 7.08FKK3 pKa = 10.78VMRR6 pKa = 11.84TLLLSLLLLATTLHH20 pKa = 6.6ASSSDD25 pKa = 3.36SGSNSTDD32 pKa = 2.96SGSNSTDD39 pKa = 2.96SGSNSTDD46 pKa = 2.96SGSNSTDD53 pKa = 2.98SSDD56 pKa = 4.92SYY58 pKa = 11.45LDD60 pKa = 3.51HH61 pKa = 7.16NKK63 pKa = 9.03TDD65 pKa = 3.67SGSGSHH71 pKa = 6.84HH72 pKa = 6.84SDD74 pKa = 2.97SGNSPDD80 pKa = 4.13SNHH83 pKa = 6.41SGSLPTDD90 pKa = 3.55SSADD94 pKa = 3.54DD95 pKa = 3.83SALSDD100 pKa = 4.48SDD102 pKa = 5.76DD103 pKa = 3.72GVCNVTHH110 pKa = 6.87CKK112 pKa = 10.04EE113 pKa = 3.88CAKK116 pKa = 10.74RR117 pKa = 11.84NVCLQCEE124 pKa = 4.28DD125 pKa = 4.65HH126 pKa = 6.9YY127 pKa = 11.63VLPPSSSTRR136 pKa = 11.84CVPVMDD142 pKa = 4.31GCLVYY147 pKa = 9.96TVNANVCQQCKK158 pKa = 9.86EE159 pKa = 4.2GYY161 pKa = 7.53TLEE164 pKa = 4.34TGGTCTGTKK173 pKa = 9.92KK174 pKa = 10.74KK175 pKa = 10.57EE176 pKa = 3.91GTQWWVWLIVALAIVVVIVIIIIIICCCCCKK207 pKa = 9.98KK208 pKa = 10.86DD209 pKa = 3.36KK210 pKa = 7.8TTPTRR215 pKa = 11.84GTSTTTSRR223 pKa = 11.84STSTNSSFSSKK234 pKa = 9.16TATGEE239 pKa = 4.05DD240 pKa = 4.36DD241 pKa = 4.55SDD243 pKa = 5.37SSDD246 pKa = 5.77DD247 pKa = 6.14DD248 pKa = 6.46DD249 pKa = 7.69DD250 pKa = 7.65DD251 pKa = 7.64DD252 pKa = 7.67DD253 pKa = 7.6DD254 pKa = 7.56DD255 pKa = 7.19DD256 pKa = 7.37DD257 pKa = 5.61EE258 pKa = 7.83DD259 pKa = 6.39SDD261 pKa = 6.34DD262 pKa = 4.81DD263 pKa = 6.15DD264 pKa = 5.15EE265 pKa = 7.02DD266 pKa = 4.48SSEE269 pKa = 4.27DD270 pKa = 3.47SSEE273 pKa = 3.98EE274 pKa = 3.84SSGDD278 pKa = 3.65EE279 pKa = 4.5EE280 pKa = 4.72EE281 pKa = 5.07DD282 pKa = 4.3SSDD285 pKa = 3.48EE286 pKa = 4.1SDD288 pKa = 3.91AEE290 pKa = 4.55GSAGTAEE297 pKa = 4.13ATVV300 pKa = 3.27
Molecular weight: 31.46 kDa
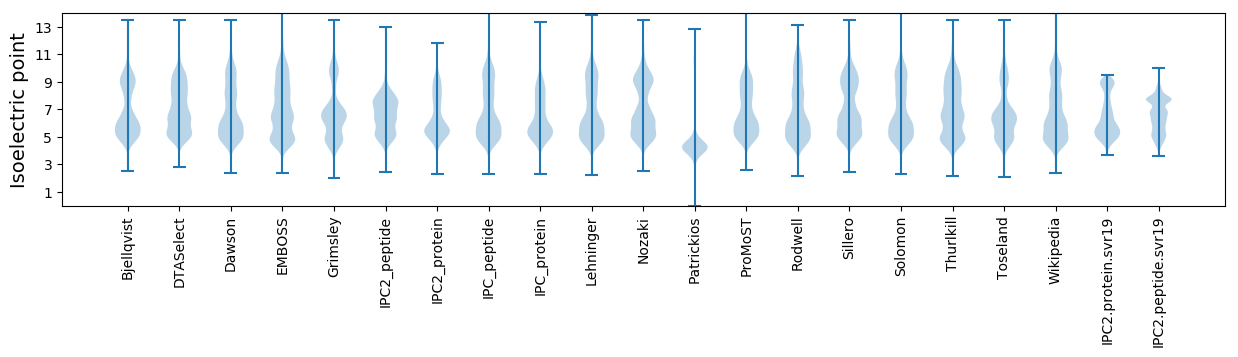
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7G2CE56|A0A7G2CE56_9TRYP Uncharacterized protein OS=Angomonas deanei OX=59799 GN=ADEAN_000471900 PE=4 SV=1
MM1 pKa = 7.31VIALATRR8 pKa = 11.84QTAMRR13 pKa = 11.84IVPATQQATTAAAPAVQWVTRR34 pKa = 11.84RR35 pKa = 11.84IALAVHH41 pKa = 6.81RR42 pKa = 11.84PTLKK46 pKa = 10.31KK47 pKa = 10.74VPAAQRR53 pKa = 11.84ATLRR57 pKa = 11.84RR58 pKa = 11.84ARR60 pKa = 11.84QATTMIARR68 pKa = 11.84AVRR71 pKa = 11.84WKK73 pKa = 8.97ITATAPVMRR82 pKa = 11.84PSPPATALVPRR93 pKa = 11.84KK94 pKa = 8.64ATLARR99 pKa = 11.84VTKK102 pKa = 10.43AALIVAKK109 pKa = 10.15AARR112 pKa = 11.84RR113 pKa = 11.84LARR116 pKa = 11.84TIALMIRR123 pKa = 11.84QATPTKK129 pKa = 10.51VRR131 pKa = 11.84AALQAKK137 pKa = 8.01LTAAPVAQQATMTKK151 pKa = 8.13TPVPHH156 pKa = 7.21RR157 pKa = 11.84VALWLATALMTLPATPKK174 pKa = 9.97KK175 pKa = 10.18VRR177 pKa = 11.84AVRR180 pKa = 11.84WKK182 pKa = 10.26IKK184 pKa = 9.49ATTPVTRR191 pKa = 11.84PSPPTTALVMQQSFLTRR208 pKa = 11.84VRR210 pKa = 11.84AVRR213 pKa = 11.84RR214 pKa = 11.84ATLPTAPVMQQSTPKK229 pKa = 10.06KK230 pKa = 8.83VRR232 pKa = 11.84AAQWVNLKK240 pKa = 9.94RR241 pKa = 11.84VPAMPRR247 pKa = 11.84ATMRR251 pKa = 11.84KK252 pKa = 7.91APAVPRR258 pKa = 11.84VITMRR263 pKa = 11.84GPAMQRR269 pKa = 11.84ATPRR273 pKa = 11.84LAAPPVRR280 pKa = 11.84PRR282 pKa = 11.84TTRR285 pKa = 11.84RR286 pKa = 11.84AALPPQRR293 pKa = 11.84HH294 pKa = 4.84NPKK297 pKa = 10.23RR298 pKa = 11.84MRR300 pKa = 11.84NDD302 pKa = 3.15LVSKK306 pKa = 10.51RR307 pKa = 11.84KK308 pKa = 9.48ADD310 pKa = 3.91RR311 pKa = 11.84PIAPLRR317 pKa = 11.84PLTEE321 pKa = 3.86QSFSSARR328 pKa = 11.84CQAALPSVTGALGKK342 pKa = 10.34
MM1 pKa = 7.31VIALATRR8 pKa = 11.84QTAMRR13 pKa = 11.84IVPATQQATTAAAPAVQWVTRR34 pKa = 11.84RR35 pKa = 11.84IALAVHH41 pKa = 6.81RR42 pKa = 11.84PTLKK46 pKa = 10.31KK47 pKa = 10.74VPAAQRR53 pKa = 11.84ATLRR57 pKa = 11.84RR58 pKa = 11.84ARR60 pKa = 11.84QATTMIARR68 pKa = 11.84AVRR71 pKa = 11.84WKK73 pKa = 8.97ITATAPVMRR82 pKa = 11.84PSPPATALVPRR93 pKa = 11.84KK94 pKa = 8.64ATLARR99 pKa = 11.84VTKK102 pKa = 10.43AALIVAKK109 pKa = 10.15AARR112 pKa = 11.84RR113 pKa = 11.84LARR116 pKa = 11.84TIALMIRR123 pKa = 11.84QATPTKK129 pKa = 10.51VRR131 pKa = 11.84AALQAKK137 pKa = 8.01LTAAPVAQQATMTKK151 pKa = 8.13TPVPHH156 pKa = 7.21RR157 pKa = 11.84VALWLATALMTLPATPKK174 pKa = 9.97KK175 pKa = 10.18VRR177 pKa = 11.84AVRR180 pKa = 11.84WKK182 pKa = 10.26IKK184 pKa = 9.49ATTPVTRR191 pKa = 11.84PSPPTTALVMQQSFLTRR208 pKa = 11.84VRR210 pKa = 11.84AVRR213 pKa = 11.84RR214 pKa = 11.84ATLPTAPVMQQSTPKK229 pKa = 10.06KK230 pKa = 8.83VRR232 pKa = 11.84AAQWVNLKK240 pKa = 9.94RR241 pKa = 11.84VPAMPRR247 pKa = 11.84ATMRR251 pKa = 11.84KK252 pKa = 7.91APAVPRR258 pKa = 11.84VITMRR263 pKa = 11.84GPAMQRR269 pKa = 11.84ATPRR273 pKa = 11.84LAAPPVRR280 pKa = 11.84PRR282 pKa = 11.84TTRR285 pKa = 11.84RR286 pKa = 11.84AALPPQRR293 pKa = 11.84HH294 pKa = 4.84NPKK297 pKa = 10.23RR298 pKa = 11.84MRR300 pKa = 11.84NDD302 pKa = 3.15LVSKK306 pKa = 10.51RR307 pKa = 11.84KK308 pKa = 9.48ADD310 pKa = 3.91RR311 pKa = 11.84PIAPLRR317 pKa = 11.84PLTEE321 pKa = 3.86QSFSSARR328 pKa = 11.84CQAALPSVTGALGKK342 pKa = 10.34
Molecular weight: 37.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4218491 |
51 |
4916 |
417.1 |
46.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.764 ± 0.04 | 1.917 ± 0.025 |
5.31 ± 0.025 | 7.451 ± 0.043 |
3.791 ± 0.018 | 5.788 ± 0.026 |
2.373 ± 0.013 | 4.086 ± 0.017 |
5.563 ± 0.026 | 9.472 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.2 ± 0.012 | 4.356 ± 0.027 |
5.033 ± 0.023 | 4.001 ± 0.026 |
6.005 ± 0.025 | 7.811 ± 0.029 |
6.05 ± 0.028 | 7.049 ± 0.022 |
0.995 ± 0.007 | 2.987 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
