
Camellia oleifera amalgavirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Amalgaviridae; unclassified Amalgaviridae
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
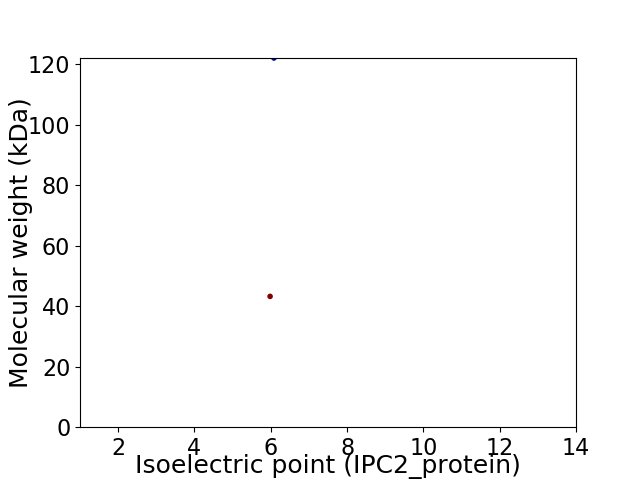
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z6JIM2|A0A2Z6JIM2_9VIRU ORF1+2p OS=Camellia oleifera amalgavirus 1 OX=2069319 GN=ORF1+ORF2 PE=4 SV=1
MM1 pKa = 7.58ANAGGNGAPRR11 pKa = 11.84PVQPLPQPPVYY22 pKa = 10.76YY23 pKa = 10.65FGQHH27 pKa = 4.96TPAEE31 pKa = 4.14NQARR35 pKa = 11.84ITEE38 pKa = 4.33AVTFLAQCGVNVGLFTVDD56 pKa = 3.14AVNQVGMTTDD66 pKa = 3.3QFIKK70 pKa = 10.35AVKK73 pKa = 9.34PLQTHH78 pKa = 6.82AEE80 pKa = 3.99QGVINPLFTEE90 pKa = 5.43AIRR93 pKa = 11.84ARR95 pKa = 11.84VFDD98 pKa = 4.32VVMEE102 pKa = 4.47LNVQQVVNVCEE113 pKa = 4.09WLKK116 pKa = 11.27SRR118 pKa = 11.84GGQNAISTVYY128 pKa = 9.16RR129 pKa = 11.84TRR131 pKa = 11.84KK132 pKa = 9.16LQAKK136 pKa = 7.73VLPGTTAADD145 pKa = 3.76VAWASVLQQQLSDD158 pKa = 3.56LSGKK162 pKa = 10.0RR163 pKa = 11.84KK164 pKa = 9.43EE165 pKa = 3.93IRR167 pKa = 11.84VEE169 pKa = 3.64KK170 pKa = 10.83DD171 pKa = 2.76EE172 pKa = 5.75AIAEE176 pKa = 4.15LRR178 pKa = 11.84RR179 pKa = 11.84EE180 pKa = 4.06IQRR183 pKa = 11.84LEE185 pKa = 3.86IEE187 pKa = 4.56KK188 pKa = 10.91GLDD191 pKa = 3.02LAAIDD196 pKa = 4.42AEE198 pKa = 4.34MVPASMYY205 pKa = 10.96NEE207 pKa = 3.9LNAAEE212 pKa = 4.18IARR215 pKa = 11.84RR216 pKa = 11.84SFNLYY221 pKa = 5.2TQRR224 pKa = 11.84AALQGVVPLPRR235 pKa = 11.84NDD237 pKa = 5.27DD238 pKa = 4.35GIKK241 pKa = 9.67MAVDD245 pKa = 3.65LFGNEE250 pKa = 3.1VRR252 pKa = 11.84QRR254 pKa = 11.84HH255 pKa = 4.63MQEE258 pKa = 4.2FVQGEE263 pKa = 4.57VVQQQLMQFLRR274 pKa = 11.84GKK276 pKa = 9.87ILEE279 pKa = 4.49LDD281 pKa = 3.76QIHH284 pKa = 6.32EE285 pKa = 4.45RR286 pKa = 11.84KK287 pKa = 9.07QASTFRR293 pKa = 11.84ALLAAVGGEE302 pKa = 4.95AITQPAVTEE311 pKa = 4.05EE312 pKa = 4.35AGDD315 pKa = 4.55DD316 pKa = 3.9GDD318 pKa = 4.16DD319 pKa = 3.73TGGEE323 pKa = 4.04AQQAGGEE330 pKa = 4.08QRR332 pKa = 11.84QDD334 pKa = 3.08APEE337 pKa = 4.11PVPARR342 pKa = 11.84VAEE345 pKa = 4.16VGEE348 pKa = 4.69GSAGDD353 pKa = 3.52QGAARR358 pKa = 11.84APPANRR364 pKa = 11.84GRR366 pKa = 11.84GRR368 pKa = 11.84RR369 pKa = 11.84GQRR372 pKa = 11.84GVQAGPRR379 pKa = 11.84KK380 pKa = 9.29SLRR383 pKa = 11.84RR384 pKa = 11.84MGRR387 pKa = 11.84GEE389 pKa = 4.19GPSATGRR396 pKa = 11.84QEE398 pKa = 3.49
MM1 pKa = 7.58ANAGGNGAPRR11 pKa = 11.84PVQPLPQPPVYY22 pKa = 10.76YY23 pKa = 10.65FGQHH27 pKa = 4.96TPAEE31 pKa = 4.14NQARR35 pKa = 11.84ITEE38 pKa = 4.33AVTFLAQCGVNVGLFTVDD56 pKa = 3.14AVNQVGMTTDD66 pKa = 3.3QFIKK70 pKa = 10.35AVKK73 pKa = 9.34PLQTHH78 pKa = 6.82AEE80 pKa = 3.99QGVINPLFTEE90 pKa = 5.43AIRR93 pKa = 11.84ARR95 pKa = 11.84VFDD98 pKa = 4.32VVMEE102 pKa = 4.47LNVQQVVNVCEE113 pKa = 4.09WLKK116 pKa = 11.27SRR118 pKa = 11.84GGQNAISTVYY128 pKa = 9.16RR129 pKa = 11.84TRR131 pKa = 11.84KK132 pKa = 9.16LQAKK136 pKa = 7.73VLPGTTAADD145 pKa = 3.76VAWASVLQQQLSDD158 pKa = 3.56LSGKK162 pKa = 10.0RR163 pKa = 11.84KK164 pKa = 9.43EE165 pKa = 3.93IRR167 pKa = 11.84VEE169 pKa = 3.64KK170 pKa = 10.83DD171 pKa = 2.76EE172 pKa = 5.75AIAEE176 pKa = 4.15LRR178 pKa = 11.84RR179 pKa = 11.84EE180 pKa = 4.06IQRR183 pKa = 11.84LEE185 pKa = 3.86IEE187 pKa = 4.56KK188 pKa = 10.91GLDD191 pKa = 3.02LAAIDD196 pKa = 4.42AEE198 pKa = 4.34MVPASMYY205 pKa = 10.96NEE207 pKa = 3.9LNAAEE212 pKa = 4.18IARR215 pKa = 11.84RR216 pKa = 11.84SFNLYY221 pKa = 5.2TQRR224 pKa = 11.84AALQGVVPLPRR235 pKa = 11.84NDD237 pKa = 5.27DD238 pKa = 4.35GIKK241 pKa = 9.67MAVDD245 pKa = 3.65LFGNEE250 pKa = 3.1VRR252 pKa = 11.84QRR254 pKa = 11.84HH255 pKa = 4.63MQEE258 pKa = 4.2FVQGEE263 pKa = 4.57VVQQQLMQFLRR274 pKa = 11.84GKK276 pKa = 9.87ILEE279 pKa = 4.49LDD281 pKa = 3.76QIHH284 pKa = 6.32EE285 pKa = 4.45RR286 pKa = 11.84KK287 pKa = 9.07QASTFRR293 pKa = 11.84ALLAAVGGEE302 pKa = 4.95AITQPAVTEE311 pKa = 4.05EE312 pKa = 4.35AGDD315 pKa = 4.55DD316 pKa = 3.9GDD318 pKa = 4.16DD319 pKa = 3.73TGGEE323 pKa = 4.04AQQAGGEE330 pKa = 4.08QRR332 pKa = 11.84QDD334 pKa = 3.08APEE337 pKa = 4.11PVPARR342 pKa = 11.84VAEE345 pKa = 4.16VGEE348 pKa = 4.69GSAGDD353 pKa = 3.52QGAARR358 pKa = 11.84APPANRR364 pKa = 11.84GRR366 pKa = 11.84GRR368 pKa = 11.84RR369 pKa = 11.84GQRR372 pKa = 11.84GVQAGPRR379 pKa = 11.84KK380 pKa = 9.29SLRR383 pKa = 11.84RR384 pKa = 11.84MGRR387 pKa = 11.84GEE389 pKa = 4.19GPSATGRR396 pKa = 11.84QEE398 pKa = 3.49
Molecular weight: 43.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z6JIM2|A0A2Z6JIM2_9VIRU ORF1+2p OS=Camellia oleifera amalgavirus 1 OX=2069319 GN=ORF1+ORF2 PE=4 SV=1
MM1 pKa = 7.58ANAGGNGAPRR11 pKa = 11.84PVQPLPQPPVYY22 pKa = 10.76YY23 pKa = 10.65FGQHH27 pKa = 4.96TPAEE31 pKa = 4.14NQARR35 pKa = 11.84ITEE38 pKa = 4.33AVTFLAQCGVNVGLFTVDD56 pKa = 3.14AVNQVGMTTDD66 pKa = 3.3QFIKK70 pKa = 10.35AVKK73 pKa = 9.34PLQTHH78 pKa = 6.82AEE80 pKa = 3.99QGVINPLFTEE90 pKa = 5.43AIRR93 pKa = 11.84ARR95 pKa = 11.84VFDD98 pKa = 4.32VVMEE102 pKa = 4.47LNVQQVVNVCEE113 pKa = 4.09WLKK116 pKa = 11.27SRR118 pKa = 11.84GGQNAISTVYY128 pKa = 9.16RR129 pKa = 11.84TRR131 pKa = 11.84KK132 pKa = 9.16LQAKK136 pKa = 7.73VLPGTTAADD145 pKa = 3.76VAWASVLQQQLSDD158 pKa = 3.56LSGKK162 pKa = 10.0RR163 pKa = 11.84KK164 pKa = 9.43EE165 pKa = 3.93IRR167 pKa = 11.84VEE169 pKa = 3.64KK170 pKa = 10.83DD171 pKa = 2.76EE172 pKa = 5.75AIAEE176 pKa = 4.15LRR178 pKa = 11.84RR179 pKa = 11.84EE180 pKa = 4.06IQRR183 pKa = 11.84LEE185 pKa = 3.86IEE187 pKa = 4.56KK188 pKa = 10.91GLDD191 pKa = 3.02LAAIDD196 pKa = 4.42AEE198 pKa = 4.34MVPASMYY205 pKa = 10.96NEE207 pKa = 3.9LNAAEE212 pKa = 4.18IARR215 pKa = 11.84RR216 pKa = 11.84SFNLYY221 pKa = 5.2TQRR224 pKa = 11.84AALQGVVPLPRR235 pKa = 11.84NDD237 pKa = 5.27DD238 pKa = 4.35GIKK241 pKa = 9.67MAVDD245 pKa = 3.65LFGNEE250 pKa = 3.1VRR252 pKa = 11.84QRR254 pKa = 11.84HH255 pKa = 4.63MQEE258 pKa = 4.2FVQGEE263 pKa = 4.57VVQQQLMQFLRR274 pKa = 11.84GKK276 pKa = 9.87ILEE279 pKa = 4.49LDD281 pKa = 3.76QIHH284 pKa = 6.32EE285 pKa = 4.45RR286 pKa = 11.84KK287 pKa = 9.04QASTFVPYY295 pKa = 9.93WLQSVVRR302 pKa = 11.84RR303 pKa = 11.84LHH305 pKa = 5.94SRR307 pKa = 11.84PLQRR311 pKa = 11.84RR312 pKa = 11.84LEE314 pKa = 4.15MMEE317 pKa = 4.57MIPVGKK323 pKa = 9.25PNKK326 pKa = 9.47PGEE329 pKa = 4.36NNVRR333 pKa = 11.84MPLSLFLPEE342 pKa = 3.99WLKK345 pKa = 11.09SEE347 pKa = 4.26KK348 pKa = 10.28GQLGIKK354 pKa = 7.15VQRR357 pKa = 11.84GLRR360 pKa = 11.84RR361 pKa = 11.84QIEE364 pKa = 4.44VVEE367 pKa = 4.82GVASEE372 pKa = 4.06EE373 pKa = 3.86FRR375 pKa = 11.84LVRR378 pKa = 11.84GKK380 pKa = 11.06VYY382 pKa = 10.7VEE384 pKa = 4.05WAAEE388 pKa = 4.17KK389 pKa = 10.86ARR391 pKa = 11.84LLPVARR397 pKa = 11.84SKK399 pKa = 11.43FEE401 pKa = 3.85TGVRR405 pKa = 11.84RR406 pKa = 11.84VIGGGEE412 pKa = 3.63MRR414 pKa = 11.84NWHH417 pKa = 6.15VASSMYY423 pKa = 10.41RR424 pKa = 11.84GGGNNADD431 pKa = 4.1ALRR434 pKa = 11.84LLSNASFMNPEE445 pKa = 4.35KK446 pKa = 10.62YY447 pKa = 10.24LHH449 pKa = 6.31EE450 pKa = 4.75CFTIEE455 pKa = 3.81TARR458 pKa = 11.84RR459 pKa = 11.84EE460 pKa = 4.15LLLTTDD466 pKa = 3.58QKK468 pKa = 11.57VPDD471 pKa = 4.29GPEE474 pKa = 3.88SVKK477 pKa = 10.33MKK479 pKa = 10.61NFNNDD484 pKa = 2.2ATAGPFLRR492 pKa = 11.84AFGIKK497 pKa = 9.73GKK499 pKa = 10.79YY500 pKa = 8.16GLKK503 pKa = 9.28TLLEE507 pKa = 3.9QVMWQFYY514 pKa = 10.69DD515 pKa = 3.76YY516 pKa = 10.85YY517 pKa = 11.43ANNEE521 pKa = 4.02VSDD524 pKa = 3.69ATLPFLTARR533 pKa = 11.84VGFRR537 pKa = 11.84TKK539 pKa = 11.1LMTEE543 pKa = 4.43RR544 pKa = 11.84KK545 pKa = 9.25ALEE548 pKa = 3.83KK549 pKa = 10.74MMVNDD554 pKa = 4.12TYY556 pKa = 11.64GRR558 pKa = 11.84AVMMLDD564 pKa = 3.68ALEE567 pKa = 4.23QAASSPLYY575 pKa = 10.59NILSSQCFHH584 pKa = 5.75GRR586 pKa = 11.84LRR588 pKa = 11.84RR589 pKa = 11.84EE590 pKa = 3.85SGFRR594 pKa = 11.84NALIRR599 pKa = 11.84ASSDD603 pKa = 2.54WHH605 pKa = 5.99FMWEE609 pKa = 4.36EE610 pKa = 3.81VQEE613 pKa = 4.05AAVIVEE619 pKa = 4.36LDD621 pKa = 2.89WSKK624 pKa = 11.03FDD626 pKa = 3.97RR627 pKa = 11.84EE628 pKa = 4.19RR629 pKa = 11.84PAEE632 pKa = 4.65DD633 pKa = 3.09IEE635 pKa = 5.05FIINVVLSCFTPKK648 pKa = 9.92TPRR651 pKa = 11.84EE652 pKa = 3.93EE653 pKa = 4.13RR654 pKa = 11.84LLRR657 pKa = 11.84AYY659 pKa = 10.49GIMMRR664 pKa = 11.84RR665 pKa = 11.84ALIEE669 pKa = 3.77RR670 pKa = 11.84LMITDD675 pKa = 3.36NAGVFGISGMVPSGSLWTGWLDD697 pKa = 3.31TALNILYY704 pKa = 9.74IRR706 pKa = 11.84AACRR710 pKa = 11.84EE711 pKa = 4.2IGVSDD716 pKa = 3.52QNSFPKK722 pKa = 10.64CAGDD726 pKa = 4.37DD727 pKa = 3.7NLTLFMRR734 pKa = 11.84DD735 pKa = 3.17PGDD738 pKa = 3.41RR739 pKa = 11.84ALNRR743 pKa = 11.84MKK745 pKa = 10.8DD746 pKa = 3.36ILNKK750 pKa = 9.28WFRR753 pKa = 11.84AGIKK757 pKa = 9.76EE758 pKa = 4.13DD759 pKa = 4.03FFIHH763 pKa = 6.53RR764 pKa = 11.84PPYY767 pKa = 9.94HH768 pKa = 6.04VSKK771 pKa = 10.42FQACFPPGTDD781 pKa = 3.58LSEE784 pKa = 4.45GTSKK788 pKa = 11.22LIDD791 pKa = 3.32EE792 pKa = 4.94AEE794 pKa = 4.09WIEE797 pKa = 3.91FHH799 pKa = 7.87DD800 pKa = 4.22EE801 pKa = 4.31LEE803 pKa = 4.4VDD805 pKa = 3.83VSRR808 pKa = 11.84GKK810 pKa = 7.87SHH812 pKa = 6.22RR813 pKa = 11.84WEE815 pKa = 3.8YY816 pKa = 10.73RR817 pKa = 11.84FKK819 pKa = 11.1GKK821 pKa = 9.56PKK823 pKa = 10.28FLSCYY828 pKa = 7.46WLEE831 pKa = 3.92NGRR834 pKa = 11.84PIRR837 pKa = 11.84PAADD841 pKa = 3.44NLEE844 pKa = 4.12KK845 pKa = 10.87LLWPEE850 pKa = 4.02GVHH853 pKa = 7.08DD854 pKa = 4.2KK855 pKa = 11.74LEE857 pKa = 4.18IYY859 pKa = 9.14EE860 pKa = 4.18AAVASMIVDD869 pKa = 3.73NPWNHH874 pKa = 6.46HH875 pKa = 5.23NVNHH879 pKa = 6.71LLMRR883 pKa = 11.84YY884 pKa = 9.93IIIQQIKK891 pKa = 9.44RR892 pKa = 11.84LGVGPANVEE901 pKa = 3.96DD902 pKa = 4.3VLFLSQFRR910 pKa = 11.84NEE912 pKa = 4.14GEE914 pKa = 4.54EE915 pKa = 3.94IPFPQVAPWRR925 pKa = 11.84KK926 pKa = 8.33FKK928 pKa = 10.7EE929 pKa = 4.07HH930 pKa = 6.03VVMEE934 pKa = 4.55EE935 pKa = 3.9YY936 pKa = 10.99NEE938 pKa = 4.05VMEE941 pKa = 4.76HH942 pKa = 6.21VEE944 pKa = 3.87NFKK947 pKa = 11.25NFLSGVTSLYY957 pKa = 10.81ARR959 pKa = 11.84RR960 pKa = 11.84AEE962 pKa = 4.3GGVDD966 pKa = 2.43AWKK969 pKa = 10.34FMEE972 pKa = 4.77IIRR975 pKa = 11.84GEE977 pKa = 4.18SHH979 pKa = 6.42VGEE982 pKa = 4.29GQFGNDD988 pKa = 4.45LIDD991 pKa = 3.33WLKK994 pKa = 10.07WMHH997 pKa = 5.45VHH999 pKa = 7.24PMTKK1003 pKa = 10.05FLRR1006 pKa = 11.84GTRR1009 pKa = 11.84MFRR1012 pKa = 11.84TPSEE1016 pKa = 4.2EE1017 pKa = 3.56IVAGDD1022 pKa = 4.1EE1023 pKa = 4.11EE1024 pKa = 4.19MDD1026 pKa = 3.76AAVEE1030 pKa = 3.98AFNVLIEE1037 pKa = 4.17RR1038 pKa = 11.84VEE1040 pKa = 4.09SEE1042 pKa = 3.97EE1043 pKa = 4.03FQDD1046 pKa = 4.11VEE1048 pKa = 5.5SFAKK1052 pKa = 9.37WVSNMLRR1059 pKa = 11.84EE1060 pKa = 4.36NVLPHH1065 pKa = 5.35QQ1066 pKa = 4.24
MM1 pKa = 7.58ANAGGNGAPRR11 pKa = 11.84PVQPLPQPPVYY22 pKa = 10.76YY23 pKa = 10.65FGQHH27 pKa = 4.96TPAEE31 pKa = 4.14NQARR35 pKa = 11.84ITEE38 pKa = 4.33AVTFLAQCGVNVGLFTVDD56 pKa = 3.14AVNQVGMTTDD66 pKa = 3.3QFIKK70 pKa = 10.35AVKK73 pKa = 9.34PLQTHH78 pKa = 6.82AEE80 pKa = 3.99QGVINPLFTEE90 pKa = 5.43AIRR93 pKa = 11.84ARR95 pKa = 11.84VFDD98 pKa = 4.32VVMEE102 pKa = 4.47LNVQQVVNVCEE113 pKa = 4.09WLKK116 pKa = 11.27SRR118 pKa = 11.84GGQNAISTVYY128 pKa = 9.16RR129 pKa = 11.84TRR131 pKa = 11.84KK132 pKa = 9.16LQAKK136 pKa = 7.73VLPGTTAADD145 pKa = 3.76VAWASVLQQQLSDD158 pKa = 3.56LSGKK162 pKa = 10.0RR163 pKa = 11.84KK164 pKa = 9.43EE165 pKa = 3.93IRR167 pKa = 11.84VEE169 pKa = 3.64KK170 pKa = 10.83DD171 pKa = 2.76EE172 pKa = 5.75AIAEE176 pKa = 4.15LRR178 pKa = 11.84RR179 pKa = 11.84EE180 pKa = 4.06IQRR183 pKa = 11.84LEE185 pKa = 3.86IEE187 pKa = 4.56KK188 pKa = 10.91GLDD191 pKa = 3.02LAAIDD196 pKa = 4.42AEE198 pKa = 4.34MVPASMYY205 pKa = 10.96NEE207 pKa = 3.9LNAAEE212 pKa = 4.18IARR215 pKa = 11.84RR216 pKa = 11.84SFNLYY221 pKa = 5.2TQRR224 pKa = 11.84AALQGVVPLPRR235 pKa = 11.84NDD237 pKa = 5.27DD238 pKa = 4.35GIKK241 pKa = 9.67MAVDD245 pKa = 3.65LFGNEE250 pKa = 3.1VRR252 pKa = 11.84QRR254 pKa = 11.84HH255 pKa = 4.63MQEE258 pKa = 4.2FVQGEE263 pKa = 4.57VVQQQLMQFLRR274 pKa = 11.84GKK276 pKa = 9.87ILEE279 pKa = 4.49LDD281 pKa = 3.76QIHH284 pKa = 6.32EE285 pKa = 4.45RR286 pKa = 11.84KK287 pKa = 9.04QASTFVPYY295 pKa = 9.93WLQSVVRR302 pKa = 11.84RR303 pKa = 11.84LHH305 pKa = 5.94SRR307 pKa = 11.84PLQRR311 pKa = 11.84RR312 pKa = 11.84LEE314 pKa = 4.15MMEE317 pKa = 4.57MIPVGKK323 pKa = 9.25PNKK326 pKa = 9.47PGEE329 pKa = 4.36NNVRR333 pKa = 11.84MPLSLFLPEE342 pKa = 3.99WLKK345 pKa = 11.09SEE347 pKa = 4.26KK348 pKa = 10.28GQLGIKK354 pKa = 7.15VQRR357 pKa = 11.84GLRR360 pKa = 11.84RR361 pKa = 11.84QIEE364 pKa = 4.44VVEE367 pKa = 4.82GVASEE372 pKa = 4.06EE373 pKa = 3.86FRR375 pKa = 11.84LVRR378 pKa = 11.84GKK380 pKa = 11.06VYY382 pKa = 10.7VEE384 pKa = 4.05WAAEE388 pKa = 4.17KK389 pKa = 10.86ARR391 pKa = 11.84LLPVARR397 pKa = 11.84SKK399 pKa = 11.43FEE401 pKa = 3.85TGVRR405 pKa = 11.84RR406 pKa = 11.84VIGGGEE412 pKa = 3.63MRR414 pKa = 11.84NWHH417 pKa = 6.15VASSMYY423 pKa = 10.41RR424 pKa = 11.84GGGNNADD431 pKa = 4.1ALRR434 pKa = 11.84LLSNASFMNPEE445 pKa = 4.35KK446 pKa = 10.62YY447 pKa = 10.24LHH449 pKa = 6.31EE450 pKa = 4.75CFTIEE455 pKa = 3.81TARR458 pKa = 11.84RR459 pKa = 11.84EE460 pKa = 4.15LLLTTDD466 pKa = 3.58QKK468 pKa = 11.57VPDD471 pKa = 4.29GPEE474 pKa = 3.88SVKK477 pKa = 10.33MKK479 pKa = 10.61NFNNDD484 pKa = 2.2ATAGPFLRR492 pKa = 11.84AFGIKK497 pKa = 9.73GKK499 pKa = 10.79YY500 pKa = 8.16GLKK503 pKa = 9.28TLLEE507 pKa = 3.9QVMWQFYY514 pKa = 10.69DD515 pKa = 3.76YY516 pKa = 10.85YY517 pKa = 11.43ANNEE521 pKa = 4.02VSDD524 pKa = 3.69ATLPFLTARR533 pKa = 11.84VGFRR537 pKa = 11.84TKK539 pKa = 11.1LMTEE543 pKa = 4.43RR544 pKa = 11.84KK545 pKa = 9.25ALEE548 pKa = 3.83KK549 pKa = 10.74MMVNDD554 pKa = 4.12TYY556 pKa = 11.64GRR558 pKa = 11.84AVMMLDD564 pKa = 3.68ALEE567 pKa = 4.23QAASSPLYY575 pKa = 10.59NILSSQCFHH584 pKa = 5.75GRR586 pKa = 11.84LRR588 pKa = 11.84RR589 pKa = 11.84EE590 pKa = 3.85SGFRR594 pKa = 11.84NALIRR599 pKa = 11.84ASSDD603 pKa = 2.54WHH605 pKa = 5.99FMWEE609 pKa = 4.36EE610 pKa = 3.81VQEE613 pKa = 4.05AAVIVEE619 pKa = 4.36LDD621 pKa = 2.89WSKK624 pKa = 11.03FDD626 pKa = 3.97RR627 pKa = 11.84EE628 pKa = 4.19RR629 pKa = 11.84PAEE632 pKa = 4.65DD633 pKa = 3.09IEE635 pKa = 5.05FIINVVLSCFTPKK648 pKa = 9.92TPRR651 pKa = 11.84EE652 pKa = 3.93EE653 pKa = 4.13RR654 pKa = 11.84LLRR657 pKa = 11.84AYY659 pKa = 10.49GIMMRR664 pKa = 11.84RR665 pKa = 11.84ALIEE669 pKa = 3.77RR670 pKa = 11.84LMITDD675 pKa = 3.36NAGVFGISGMVPSGSLWTGWLDD697 pKa = 3.31TALNILYY704 pKa = 9.74IRR706 pKa = 11.84AACRR710 pKa = 11.84EE711 pKa = 4.2IGVSDD716 pKa = 3.52QNSFPKK722 pKa = 10.64CAGDD726 pKa = 4.37DD727 pKa = 3.7NLTLFMRR734 pKa = 11.84DD735 pKa = 3.17PGDD738 pKa = 3.41RR739 pKa = 11.84ALNRR743 pKa = 11.84MKK745 pKa = 10.8DD746 pKa = 3.36ILNKK750 pKa = 9.28WFRR753 pKa = 11.84AGIKK757 pKa = 9.76EE758 pKa = 4.13DD759 pKa = 4.03FFIHH763 pKa = 6.53RR764 pKa = 11.84PPYY767 pKa = 9.94HH768 pKa = 6.04VSKK771 pKa = 10.42FQACFPPGTDD781 pKa = 3.58LSEE784 pKa = 4.45GTSKK788 pKa = 11.22LIDD791 pKa = 3.32EE792 pKa = 4.94AEE794 pKa = 4.09WIEE797 pKa = 3.91FHH799 pKa = 7.87DD800 pKa = 4.22EE801 pKa = 4.31LEE803 pKa = 4.4VDD805 pKa = 3.83VSRR808 pKa = 11.84GKK810 pKa = 7.87SHH812 pKa = 6.22RR813 pKa = 11.84WEE815 pKa = 3.8YY816 pKa = 10.73RR817 pKa = 11.84FKK819 pKa = 11.1GKK821 pKa = 9.56PKK823 pKa = 10.28FLSCYY828 pKa = 7.46WLEE831 pKa = 3.92NGRR834 pKa = 11.84PIRR837 pKa = 11.84PAADD841 pKa = 3.44NLEE844 pKa = 4.12KK845 pKa = 10.87LLWPEE850 pKa = 4.02GVHH853 pKa = 7.08DD854 pKa = 4.2KK855 pKa = 11.74LEE857 pKa = 4.18IYY859 pKa = 9.14EE860 pKa = 4.18AAVASMIVDD869 pKa = 3.73NPWNHH874 pKa = 6.46HH875 pKa = 5.23NVNHH879 pKa = 6.71LLMRR883 pKa = 11.84YY884 pKa = 9.93IIIQQIKK891 pKa = 9.44RR892 pKa = 11.84LGVGPANVEE901 pKa = 3.96DD902 pKa = 4.3VLFLSQFRR910 pKa = 11.84NEE912 pKa = 4.14GEE914 pKa = 4.54EE915 pKa = 3.94IPFPQVAPWRR925 pKa = 11.84KK926 pKa = 8.33FKK928 pKa = 10.7EE929 pKa = 4.07HH930 pKa = 6.03VVMEE934 pKa = 4.55EE935 pKa = 3.9YY936 pKa = 10.99NEE938 pKa = 4.05VMEE941 pKa = 4.76HH942 pKa = 6.21VEE944 pKa = 3.87NFKK947 pKa = 11.25NFLSGVTSLYY957 pKa = 10.81ARR959 pKa = 11.84RR960 pKa = 11.84AEE962 pKa = 4.3GGVDD966 pKa = 2.43AWKK969 pKa = 10.34FMEE972 pKa = 4.77IIRR975 pKa = 11.84GEE977 pKa = 4.18SHH979 pKa = 6.42VGEE982 pKa = 4.29GQFGNDD988 pKa = 4.45LIDD991 pKa = 3.33WLKK994 pKa = 10.07WMHH997 pKa = 5.45VHH999 pKa = 7.24PMTKK1003 pKa = 10.05FLRR1006 pKa = 11.84GTRR1009 pKa = 11.84MFRR1012 pKa = 11.84TPSEE1016 pKa = 4.2EE1017 pKa = 3.56IVAGDD1022 pKa = 4.1EE1023 pKa = 4.11EE1024 pKa = 4.19MDD1026 pKa = 3.76AAVEE1030 pKa = 3.98AFNVLIEE1037 pKa = 4.17RR1038 pKa = 11.84VEE1040 pKa = 4.09SEE1042 pKa = 3.97EE1043 pKa = 4.03FQDD1046 pKa = 4.11VEE1048 pKa = 5.5SFAKK1052 pKa = 9.37WVSNMLRR1059 pKa = 11.84EE1060 pKa = 4.36NVLPHH1065 pKa = 5.35QQ1066 pKa = 4.24
Molecular weight: 122.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1464 |
398 |
1066 |
732.0 |
82.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.016 ± 1.586 | 0.751 ± 0.12 |
4.44 ± 0.04 | 8.538 ± 0.361 |
4.303 ± 0.741 | 7.377 ± 1.166 |
1.844 ± 0.404 | 4.508 ± 0.356 |
4.44 ± 0.565 | 8.402 ± 0.658 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.279 ± 0.49 | 4.577 ± 0.389 |
4.781 ± 0.238 | 5.806 ± 1.68 |
7.923 ± 0.298 | 4.098 ± 0.642 |
3.825 ± 0.336 | 8.402 ± 0.31 |
1.708 ± 0.58 | 1.981 ± 0.349 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
