
Pararhodobacter oceanensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Pararhodobacter
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
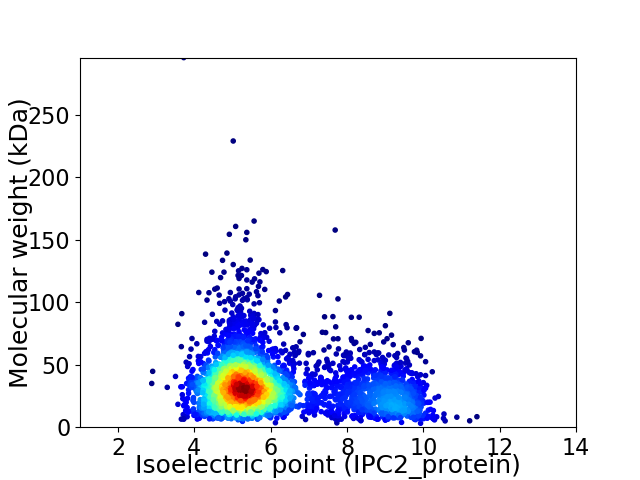
Virtual 2D-PAGE plot for 3661 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T8HVW2|A0A2T8HVW2_9RHOB DsbE family thiol:disulfide interchange protein OS=Pararhodobacter oceanensis OX=2172121 GN=DDE20_05495 PE=3 SV=1
MM1 pKa = 7.48TKK3 pKa = 10.41FISGRR8 pKa = 11.84FKK10 pKa = 11.06GVGGGVAAVAALSLPVLPITAAAQSEE36 pKa = 4.6GAAGGFAIEE45 pKa = 4.71VGPSIDD51 pKa = 3.34PYY53 pKa = 11.03LPVRR57 pKa = 11.84VGARR61 pKa = 11.84VFGEE65 pKa = 3.86ASEE68 pKa = 4.22PASGFVRR75 pKa = 11.84GCQGFVLPEE84 pKa = 3.55SAGAVFEE91 pKa = 4.48VTEE94 pKa = 4.37EE95 pKa = 3.99MATLAFTGAAEE106 pKa = 4.45GLASMVLGTPDD117 pKa = 5.11GLFQCALADD126 pKa = 3.78EE127 pKa = 5.15RR128 pKa = 11.84GLAVVNMAGVAPGRR142 pKa = 11.84YY143 pKa = 9.08SVWLGAAEE151 pKa = 4.45GAQIDD156 pKa = 3.88ARR158 pKa = 11.84LFASDD163 pKa = 3.9SVISAIEE170 pKa = 3.83IFGLDD175 pKa = 3.74MAHH178 pKa = 6.35MGEE181 pKa = 4.33PRR183 pKa = 11.84VGRR186 pKa = 11.84HH187 pKa = 4.95VFSASLEE194 pKa = 4.21SGRR197 pKa = 11.84QEE199 pKa = 4.07LASGAIVSAVSEE211 pKa = 4.32MRR213 pKa = 11.84PLSPDD218 pKa = 3.11YY219 pKa = 11.29CPGYY223 pKa = 9.59TNFDD227 pKa = 3.36AADD230 pKa = 3.45AVLTLDD236 pKa = 4.23AEE238 pKa = 4.35IDD240 pKa = 3.76RR241 pKa = 11.84FSVFATSQRR250 pKa = 11.84DD251 pKa = 3.18LTMAVVQPDD260 pKa = 3.78GAVICNDD267 pKa = 3.86DD268 pKa = 4.13AFEE271 pKa = 4.03LHH273 pKa = 6.95PGVVIDD279 pKa = 4.37RR280 pKa = 11.84AQAGDD285 pKa = 3.33YY286 pKa = 10.43HH287 pKa = 7.69VFVGAYY293 pKa = 8.98SQGGTASFDD302 pKa = 3.6LFASEE307 pKa = 4.87GGPAFSNAILDD318 pKa = 3.91LDD320 pKa = 3.95AAPRR324 pKa = 11.84AGSALFDD331 pKa = 3.66MDD333 pKa = 5.1AAGQGQLLAAGQVSAVDD350 pKa = 3.79PMASLATGNYY360 pKa = 8.77CPGYY364 pKa = 8.84TDD366 pKa = 3.7ISAPDD371 pKa = 3.71FVMTMDD377 pKa = 3.56QAQPMISLYY386 pKa = 10.82ARR388 pKa = 11.84SEE390 pKa = 4.13TDD392 pKa = 2.82LVLAVRR398 pKa = 11.84APDD401 pKa = 3.98GTWLCNDD408 pKa = 4.9DD409 pKa = 5.21NYY411 pKa = 11.12QLNPGVTLQEE421 pKa = 4.16AQAGDD426 pKa = 3.39YY427 pKa = 10.54HH428 pKa = 7.55VFVGSYY434 pKa = 8.21HH435 pKa = 5.71TGAMGGYY442 pKa = 9.46NLYY445 pKa = 11.19ASMGAPNWQEE455 pKa = 3.9TQGGGGSGLGVLDD468 pKa = 3.75ITAEE472 pKa = 3.99PAIGRR477 pKa = 11.84IGFGPDD483 pKa = 2.99TQIDD487 pKa = 3.54PRR489 pKa = 11.84IIFDD493 pKa = 3.67IEE495 pKa = 3.94ASQTEE500 pKa = 4.47AFGMGPGCAGFITPEE515 pKa = 4.09RR516 pKa = 11.84PDD518 pKa = 3.46LVINAEE524 pKa = 4.09AGLPQLMVYY533 pKa = 8.39MASEE537 pKa = 4.22ADD539 pKa = 3.43GTLVISGPDD548 pKa = 3.45GQIYY552 pKa = 10.66CNDD555 pKa = 4.11DD556 pKa = 3.66FEE558 pKa = 4.53QLNPGVMIPNAPAGDD573 pKa = 3.66YY574 pKa = 10.99SVFAGTYY581 pKa = 9.48SGTGGLATLGVTIAAPQWVMDD602 pKa = 4.34RR603 pKa = 11.84EE604 pKa = 4.4HH605 pKa = 6.82
MM1 pKa = 7.48TKK3 pKa = 10.41FISGRR8 pKa = 11.84FKK10 pKa = 11.06GVGGGVAAVAALSLPVLPITAAAQSEE36 pKa = 4.6GAAGGFAIEE45 pKa = 4.71VGPSIDD51 pKa = 3.34PYY53 pKa = 11.03LPVRR57 pKa = 11.84VGARR61 pKa = 11.84VFGEE65 pKa = 3.86ASEE68 pKa = 4.22PASGFVRR75 pKa = 11.84GCQGFVLPEE84 pKa = 3.55SAGAVFEE91 pKa = 4.48VTEE94 pKa = 4.37EE95 pKa = 3.99MATLAFTGAAEE106 pKa = 4.45GLASMVLGTPDD117 pKa = 5.11GLFQCALADD126 pKa = 3.78EE127 pKa = 5.15RR128 pKa = 11.84GLAVVNMAGVAPGRR142 pKa = 11.84YY143 pKa = 9.08SVWLGAAEE151 pKa = 4.45GAQIDD156 pKa = 3.88ARR158 pKa = 11.84LFASDD163 pKa = 3.9SVISAIEE170 pKa = 3.83IFGLDD175 pKa = 3.74MAHH178 pKa = 6.35MGEE181 pKa = 4.33PRR183 pKa = 11.84VGRR186 pKa = 11.84HH187 pKa = 4.95VFSASLEE194 pKa = 4.21SGRR197 pKa = 11.84QEE199 pKa = 4.07LASGAIVSAVSEE211 pKa = 4.32MRR213 pKa = 11.84PLSPDD218 pKa = 3.11YY219 pKa = 11.29CPGYY223 pKa = 9.59TNFDD227 pKa = 3.36AADD230 pKa = 3.45AVLTLDD236 pKa = 4.23AEE238 pKa = 4.35IDD240 pKa = 3.76RR241 pKa = 11.84FSVFATSQRR250 pKa = 11.84DD251 pKa = 3.18LTMAVVQPDD260 pKa = 3.78GAVICNDD267 pKa = 3.86DD268 pKa = 4.13AFEE271 pKa = 4.03LHH273 pKa = 6.95PGVVIDD279 pKa = 4.37RR280 pKa = 11.84AQAGDD285 pKa = 3.33YY286 pKa = 10.43HH287 pKa = 7.69VFVGAYY293 pKa = 8.98SQGGTASFDD302 pKa = 3.6LFASEE307 pKa = 4.87GGPAFSNAILDD318 pKa = 3.91LDD320 pKa = 3.95AAPRR324 pKa = 11.84AGSALFDD331 pKa = 3.66MDD333 pKa = 5.1AAGQGQLLAAGQVSAVDD350 pKa = 3.79PMASLATGNYY360 pKa = 8.77CPGYY364 pKa = 8.84TDD366 pKa = 3.7ISAPDD371 pKa = 3.71FVMTMDD377 pKa = 3.56QAQPMISLYY386 pKa = 10.82ARR388 pKa = 11.84SEE390 pKa = 4.13TDD392 pKa = 2.82LVLAVRR398 pKa = 11.84APDD401 pKa = 3.98GTWLCNDD408 pKa = 4.9DD409 pKa = 5.21NYY411 pKa = 11.12QLNPGVTLQEE421 pKa = 4.16AQAGDD426 pKa = 3.39YY427 pKa = 10.54HH428 pKa = 7.55VFVGSYY434 pKa = 8.21HH435 pKa = 5.71TGAMGGYY442 pKa = 9.46NLYY445 pKa = 11.19ASMGAPNWQEE455 pKa = 3.9TQGGGGSGLGVLDD468 pKa = 3.75ITAEE472 pKa = 3.99PAIGRR477 pKa = 11.84IGFGPDD483 pKa = 2.99TQIDD487 pKa = 3.54PRR489 pKa = 11.84IIFDD493 pKa = 3.67IEE495 pKa = 3.94ASQTEE500 pKa = 4.47AFGMGPGCAGFITPEE515 pKa = 4.09RR516 pKa = 11.84PDD518 pKa = 3.46LVINAEE524 pKa = 4.09AGLPQLMVYY533 pKa = 8.39MASEE537 pKa = 4.22ADD539 pKa = 3.43GTLVISGPDD548 pKa = 3.45GQIYY552 pKa = 10.66CNDD555 pKa = 4.11DD556 pKa = 3.66FEE558 pKa = 4.53QLNPGVMIPNAPAGDD573 pKa = 3.66YY574 pKa = 10.99SVFAGTYY581 pKa = 9.48SGTGGLATLGVTIAAPQWVMDD602 pKa = 4.34RR603 pKa = 11.84EE604 pKa = 4.4HH605 pKa = 6.82
Molecular weight: 62.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T8HVS9|A0A2T8HVS9_9RHOB DNA-directed RNA polymerase subunit beta' OS=Pararhodobacter oceanensis OX=2172121 GN=rpoC PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.35GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.35GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1181261 |
29 |
2887 |
322.7 |
34.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.982 ± 0.058 | 0.881 ± 0.012 |
5.694 ± 0.031 | 5.848 ± 0.035 |
3.654 ± 0.027 | 8.666 ± 0.036 |
2.146 ± 0.017 | 5.208 ± 0.025 |
2.622 ± 0.032 | 10.307 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.826 ± 0.018 | 2.511 ± 0.022 |
5.223 ± 0.029 | 3.382 ± 0.023 |
6.873 ± 0.035 | 5.18 ± 0.028 |
5.348 ± 0.035 | 7.102 ± 0.037 |
1.386 ± 0.018 | 2.161 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
