
Sewage-associated circular DNA virus-6
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.21
Get precalculated fractions of proteins
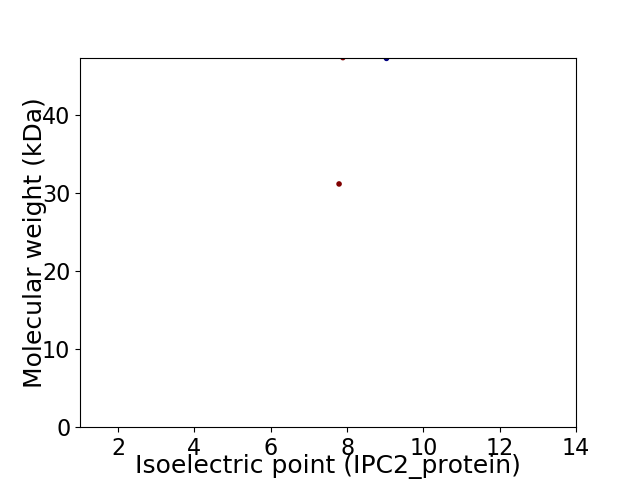
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
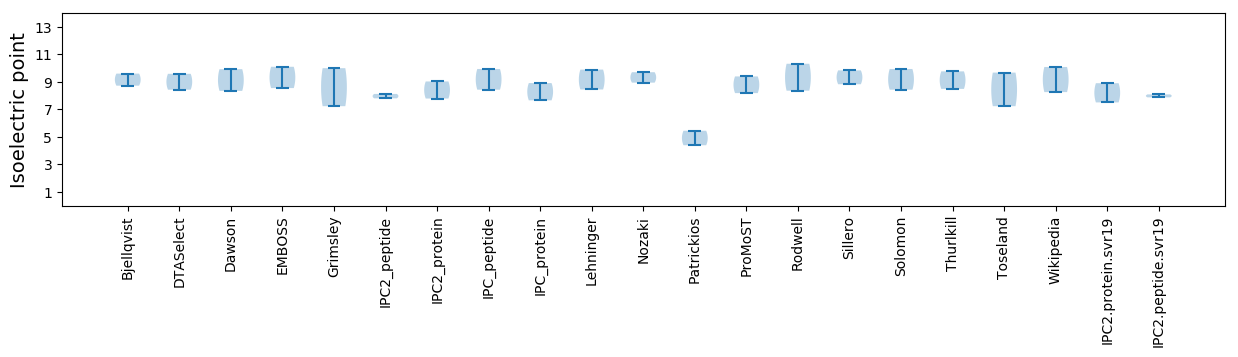
Protein with the lowest isoelectric point:
>tr|A0A075J182|A0A075J182_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-6 OX=1519395 PE=4 SV=1
MM1 pKa = 7.72GEE3 pKa = 3.82ARR5 pKa = 11.84RR6 pKa = 11.84QGVYY10 pKa = 9.01WLLTIPHH17 pKa = 7.22ADD19 pKa = 3.06WTPYY23 pKa = 10.26LPPGITWIRR32 pKa = 11.84GQLEE36 pKa = 4.31SGSDD40 pKa = 2.91TGYY43 pKa = 10.8LHH45 pKa = 5.96WQIIVGCSKK54 pKa = 10.44KK55 pKa = 10.32KK56 pKa = 9.55SLRR59 pKa = 11.84GIQEE63 pKa = 3.58IFGRR67 pKa = 11.84TCHH70 pKa = 6.65AEE72 pKa = 3.9LGRR75 pKa = 11.84SEE77 pKa = 4.61AAAAYY82 pKa = 8.71VWKK85 pKa = 10.47EE86 pKa = 3.68NTRR89 pKa = 11.84VEE91 pKa = 4.35GTQFEE96 pKa = 4.64LGKK99 pKa = 10.79KK100 pKa = 8.68PFKK103 pKa = 10.59RR104 pKa = 11.84NCATDD109 pKa = 3.01WDD111 pKa = 5.31LIWEE115 pKa = 4.31SAQRR119 pKa = 11.84GDD121 pKa = 4.48FMSIPADD128 pKa = 3.38IRR130 pKa = 11.84IHH132 pKa = 6.14SYY134 pKa = 8.66RR135 pKa = 11.84TLRR138 pKa = 11.84AISADD143 pKa = 3.42YY144 pKa = 10.63AKK146 pKa = 9.89PIEE149 pKa = 4.21MEE151 pKa = 3.87RR152 pKa = 11.84TCYY155 pKa = 10.75VFVGPTGTGKK165 pKa = 9.99SRR167 pKa = 11.84RR168 pKa = 11.84AWSEE172 pKa = 3.55SGMEE176 pKa = 4.55AFCKK180 pKa = 10.59DD181 pKa = 3.58PNSKK185 pKa = 9.74FWCGYY190 pKa = 7.91SGQSRR195 pKa = 11.84VVIDD199 pKa = 3.65EE200 pKa = 3.67FRR202 pKa = 11.84GRR204 pKa = 11.84IDD206 pKa = 3.22VSHH209 pKa = 7.5LLRR212 pKa = 11.84WLDD215 pKa = 3.64RR216 pKa = 11.84YY217 pKa = 9.52PCNVEE222 pKa = 3.74IKK224 pKa = 10.13GSSVPLCAEE233 pKa = 4.31SFWITSNLDD242 pKa = 2.7VDD244 pKa = 3.38QWYY247 pKa = 9.46PEE249 pKa = 4.17LDD251 pKa = 3.98EE252 pKa = 4.66PTLAALKK259 pKa = 10.14RR260 pKa = 11.84RR261 pKa = 11.84LKK263 pKa = 8.97ITHH266 pKa = 6.4FRR268 pKa = 11.84TLQQ271 pKa = 3.2
MM1 pKa = 7.72GEE3 pKa = 3.82ARR5 pKa = 11.84RR6 pKa = 11.84QGVYY10 pKa = 9.01WLLTIPHH17 pKa = 7.22ADD19 pKa = 3.06WTPYY23 pKa = 10.26LPPGITWIRR32 pKa = 11.84GQLEE36 pKa = 4.31SGSDD40 pKa = 2.91TGYY43 pKa = 10.8LHH45 pKa = 5.96WQIIVGCSKK54 pKa = 10.44KK55 pKa = 10.32KK56 pKa = 9.55SLRR59 pKa = 11.84GIQEE63 pKa = 3.58IFGRR67 pKa = 11.84TCHH70 pKa = 6.65AEE72 pKa = 3.9LGRR75 pKa = 11.84SEE77 pKa = 4.61AAAAYY82 pKa = 8.71VWKK85 pKa = 10.47EE86 pKa = 3.68NTRR89 pKa = 11.84VEE91 pKa = 4.35GTQFEE96 pKa = 4.64LGKK99 pKa = 10.79KK100 pKa = 8.68PFKK103 pKa = 10.59RR104 pKa = 11.84NCATDD109 pKa = 3.01WDD111 pKa = 5.31LIWEE115 pKa = 4.31SAQRR119 pKa = 11.84GDD121 pKa = 4.48FMSIPADD128 pKa = 3.38IRR130 pKa = 11.84IHH132 pKa = 6.14SYY134 pKa = 8.66RR135 pKa = 11.84TLRR138 pKa = 11.84AISADD143 pKa = 3.42YY144 pKa = 10.63AKK146 pKa = 9.89PIEE149 pKa = 4.21MEE151 pKa = 3.87RR152 pKa = 11.84TCYY155 pKa = 10.75VFVGPTGTGKK165 pKa = 9.99SRR167 pKa = 11.84RR168 pKa = 11.84AWSEE172 pKa = 3.55SGMEE176 pKa = 4.55AFCKK180 pKa = 10.59DD181 pKa = 3.58PNSKK185 pKa = 9.74FWCGYY190 pKa = 7.91SGQSRR195 pKa = 11.84VVIDD199 pKa = 3.65EE200 pKa = 3.67FRR202 pKa = 11.84GRR204 pKa = 11.84IDD206 pKa = 3.22VSHH209 pKa = 7.5LLRR212 pKa = 11.84WLDD215 pKa = 3.64RR216 pKa = 11.84YY217 pKa = 9.52PCNVEE222 pKa = 3.74IKK224 pKa = 10.13GSSVPLCAEE233 pKa = 4.31SFWITSNLDD242 pKa = 2.7VDD244 pKa = 3.38QWYY247 pKa = 9.46PEE249 pKa = 4.17LDD251 pKa = 3.98EE252 pKa = 4.66PTLAALKK259 pKa = 10.14RR260 pKa = 11.84RR261 pKa = 11.84LKK263 pKa = 8.97ITHH266 pKa = 6.4FRR268 pKa = 11.84TLQQ271 pKa = 3.2
Molecular weight: 31.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075J182|A0A075J182_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-6 OX=1519395 PE=4 SV=1
MM1 pKa = 6.12VTIRR5 pKa = 11.84EE6 pKa = 4.25LAGGLSGGALGYY18 pKa = 8.29ITHH21 pKa = 7.17GARR24 pKa = 11.84GAIKK28 pKa = 10.13GASAGYY34 pKa = 10.34ALARR38 pKa = 11.84NKK40 pKa = 10.22RR41 pKa = 11.84KK42 pKa = 10.44AEE44 pKa = 3.95DD45 pKa = 3.53DD46 pKa = 3.3MAGNRR51 pKa = 11.84KK52 pKa = 8.22YY53 pKa = 11.24VKK55 pKa = 9.48TKK57 pKa = 10.52HH58 pKa = 6.47PYY60 pKa = 8.4TKK62 pKa = 10.55VNKK65 pKa = 9.73LSKK68 pKa = 10.47KK69 pKa = 9.59YY70 pKa = 9.98GLHH73 pKa = 6.2RR74 pKa = 11.84ARR76 pKa = 11.84NISGGGPHH84 pKa = 7.82PYY86 pKa = 10.66NKK88 pKa = 9.91VGSNASAVKK97 pKa = 10.47LKK99 pKa = 10.35RR100 pKa = 11.84KK101 pKa = 9.95KK102 pKa = 10.33GVEE105 pKa = 3.29IKK107 pKa = 9.8KK108 pKa = 9.4RR109 pKa = 11.84KK110 pKa = 8.36RR111 pKa = 11.84VRR113 pKa = 11.84VSKK116 pKa = 10.8LFEE119 pKa = 4.52KK120 pKa = 10.47KK121 pKa = 10.0VKK123 pKa = 9.69QALEE127 pKa = 4.17AEE129 pKa = 4.97DD130 pKa = 4.86IEE132 pKa = 4.6GTYY135 pKa = 10.95LKK137 pKa = 9.49VTYY140 pKa = 11.01ARR142 pKa = 11.84MLPPASDD149 pKa = 3.3TSQAVIDD156 pKa = 3.96FANAFGPMEE165 pKa = 4.44FVDD168 pKa = 4.05AADD171 pKa = 3.5VLFNDD176 pKa = 4.02QVPVEE181 pKa = 4.92APTFPNNDD189 pKa = 2.92WSNNWIRR196 pKa = 11.84KK197 pKa = 7.21DD198 pKa = 3.49TVINSTMQLEE208 pKa = 4.69MKK210 pKa = 10.59NMSQRR215 pKa = 11.84TYY217 pKa = 7.81TLKK220 pKa = 10.35IYY222 pKa = 8.64TCQPKK227 pKa = 9.63VVPTDD232 pKa = 3.46AAPNDD237 pKa = 4.1ALADD241 pKa = 3.72WSTGMIMSTSFGANPLSNSPSTLYY265 pKa = 10.54CIPTDD270 pKa = 3.22APQFNRR276 pKa = 11.84FWKK279 pKa = 9.16AQCEE283 pKa = 4.42TVVLQPGQSHH293 pKa = 6.38TYY295 pKa = 10.03FLQGPNDD302 pKa = 3.45YY303 pKa = 11.36VLDD306 pKa = 3.99YY307 pKa = 11.26KK308 pKa = 10.62KK309 pKa = 11.04LQMKK313 pKa = 10.26NIAAAPNWLIPYY325 pKa = 10.27AKK327 pKa = 9.07FTRR330 pKa = 11.84NVFVVGHH337 pKa = 6.41TDD339 pKa = 3.58LVTSTLAGAGRR350 pKa = 11.84FPSGGVGQGGIVFEE364 pKa = 4.07RR365 pKa = 11.84RR366 pKa = 11.84MFYY369 pKa = 10.96KK370 pKa = 9.82MKK372 pKa = 10.06CPASAGFQYY381 pKa = 8.45PTGAYY386 pKa = 8.89PVGDD390 pKa = 3.9TQQLDD395 pKa = 3.46QKK397 pKa = 10.83IPAKK401 pKa = 10.16VIKK404 pKa = 9.75FFPIGAAGSTEE415 pKa = 4.22DD416 pKa = 3.84VLEE419 pKa = 4.38EE420 pKa = 4.17NPLSIINPLDD430 pKa = 3.17
MM1 pKa = 6.12VTIRR5 pKa = 11.84EE6 pKa = 4.25LAGGLSGGALGYY18 pKa = 8.29ITHH21 pKa = 7.17GARR24 pKa = 11.84GAIKK28 pKa = 10.13GASAGYY34 pKa = 10.34ALARR38 pKa = 11.84NKK40 pKa = 10.22RR41 pKa = 11.84KK42 pKa = 10.44AEE44 pKa = 3.95DD45 pKa = 3.53DD46 pKa = 3.3MAGNRR51 pKa = 11.84KK52 pKa = 8.22YY53 pKa = 11.24VKK55 pKa = 9.48TKK57 pKa = 10.52HH58 pKa = 6.47PYY60 pKa = 8.4TKK62 pKa = 10.55VNKK65 pKa = 9.73LSKK68 pKa = 10.47KK69 pKa = 9.59YY70 pKa = 9.98GLHH73 pKa = 6.2RR74 pKa = 11.84ARR76 pKa = 11.84NISGGGPHH84 pKa = 7.82PYY86 pKa = 10.66NKK88 pKa = 9.91VGSNASAVKK97 pKa = 10.47LKK99 pKa = 10.35RR100 pKa = 11.84KK101 pKa = 9.95KK102 pKa = 10.33GVEE105 pKa = 3.29IKK107 pKa = 9.8KK108 pKa = 9.4RR109 pKa = 11.84KK110 pKa = 8.36RR111 pKa = 11.84VRR113 pKa = 11.84VSKK116 pKa = 10.8LFEE119 pKa = 4.52KK120 pKa = 10.47KK121 pKa = 10.0VKK123 pKa = 9.69QALEE127 pKa = 4.17AEE129 pKa = 4.97DD130 pKa = 4.86IEE132 pKa = 4.6GTYY135 pKa = 10.95LKK137 pKa = 9.49VTYY140 pKa = 11.01ARR142 pKa = 11.84MLPPASDD149 pKa = 3.3TSQAVIDD156 pKa = 3.96FANAFGPMEE165 pKa = 4.44FVDD168 pKa = 4.05AADD171 pKa = 3.5VLFNDD176 pKa = 4.02QVPVEE181 pKa = 4.92APTFPNNDD189 pKa = 2.92WSNNWIRR196 pKa = 11.84KK197 pKa = 7.21DD198 pKa = 3.49TVINSTMQLEE208 pKa = 4.69MKK210 pKa = 10.59NMSQRR215 pKa = 11.84TYY217 pKa = 7.81TLKK220 pKa = 10.35IYY222 pKa = 8.64TCQPKK227 pKa = 9.63VVPTDD232 pKa = 3.46AAPNDD237 pKa = 4.1ALADD241 pKa = 3.72WSTGMIMSTSFGANPLSNSPSTLYY265 pKa = 10.54CIPTDD270 pKa = 3.22APQFNRR276 pKa = 11.84FWKK279 pKa = 9.16AQCEE283 pKa = 4.42TVVLQPGQSHH293 pKa = 6.38TYY295 pKa = 10.03FLQGPNDD302 pKa = 3.45YY303 pKa = 11.36VLDD306 pKa = 3.99YY307 pKa = 11.26KK308 pKa = 10.62KK309 pKa = 11.04LQMKK313 pKa = 10.26NIAAAPNWLIPYY325 pKa = 10.27AKK327 pKa = 9.07FTRR330 pKa = 11.84NVFVVGHH337 pKa = 6.41TDD339 pKa = 3.58LVTSTLAGAGRR350 pKa = 11.84FPSGGVGQGGIVFEE364 pKa = 4.07RR365 pKa = 11.84RR366 pKa = 11.84MFYY369 pKa = 10.96KK370 pKa = 9.82MKK372 pKa = 10.06CPASAGFQYY381 pKa = 8.45PTGAYY386 pKa = 8.89PVGDD390 pKa = 3.9TQQLDD395 pKa = 3.46QKK397 pKa = 10.83IPAKK401 pKa = 10.16VIKK404 pKa = 9.75FFPIGAAGSTEE415 pKa = 4.22DD416 pKa = 3.84VLEE419 pKa = 4.38EE420 pKa = 4.17NPLSIINPLDD430 pKa = 3.17
Molecular weight: 47.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
701 |
271 |
430 |
350.5 |
39.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.559 ± 1.089 | 1.712 ± 0.705 |
4.993 ± 0.098 | 4.708 ± 1.099 |
3.994 ± 0.173 | 7.989 ± 0.136 |
1.712 ± 0.285 | 5.421 ± 0.694 |
7.275 ± 1.199 | 6.847 ± 0.303 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.282 ± 0.458 | 4.137 ± 1.302 |
5.849 ± 0.598 | 3.709 ± 0.22 |
5.991 ± 1.418 | 6.277 ± 0.627 |
6.134 ± 0.131 | 5.991 ± 0.888 |
2.425 ± 1.138 | 3.994 ± 0.173 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
