
Bifidobacterium phage BadAargau2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; Badaztecvirus; Bifidobacterium virus BadAargau2
Average proteome isoelectric point is 5.61
Get precalculated fractions of proteins
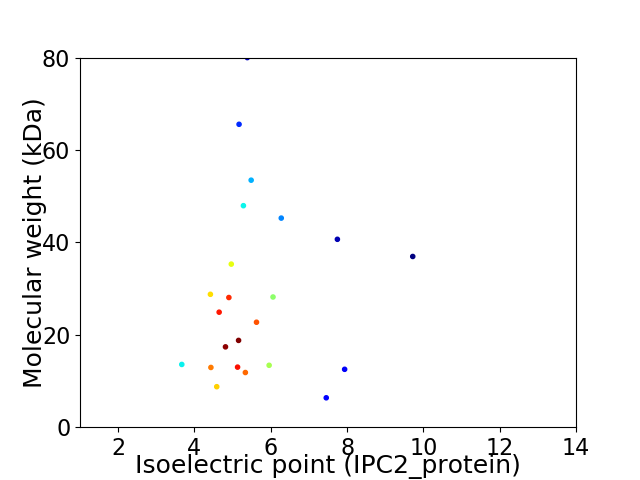
Virtual 2D-PAGE plot for 23 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G6Y0H1|A0A6G6Y0H1_9CAUD Uncharacterized protein OS=Bifidobacterium phage BadAargau2 OX=2713242 GN=BAAR0010003c01_00012 PE=4 SV=1
MM1 pKa = 7.3QAIKK5 pKa = 10.92DD6 pKa = 3.99EE7 pKa = 4.56LTEE10 pKa = 4.4SMDD13 pKa = 4.3DD14 pKa = 3.73AVEE17 pKa = 3.95LASAVVADD25 pKa = 4.48WSGHH29 pKa = 4.15MDD31 pKa = 3.14VTVRR35 pKa = 11.84PEE37 pKa = 3.46YY38 pKa = 9.26MAIVVTCGPVRR49 pKa = 11.84LWIKK53 pKa = 10.61APYY56 pKa = 9.49GSSLITAYY64 pKa = 10.59FPVSGYY70 pKa = 11.16AEE72 pKa = 4.16SCDD75 pKa = 3.22IEE77 pKa = 4.71HH78 pKa = 6.58EE79 pKa = 4.42NKK81 pKa = 10.35LGEE84 pKa = 4.68YY85 pKa = 8.71FTMIQSSDD93 pKa = 3.7MEE95 pKa = 4.1QLRR98 pKa = 11.84YY99 pKa = 10.61DD100 pKa = 3.07IMNWIRR106 pKa = 11.84VRR108 pKa = 11.84LVMSCLL114 pKa = 3.45
MM1 pKa = 7.3QAIKK5 pKa = 10.92DD6 pKa = 3.99EE7 pKa = 4.56LTEE10 pKa = 4.4SMDD13 pKa = 4.3DD14 pKa = 3.73AVEE17 pKa = 3.95LASAVVADD25 pKa = 4.48WSGHH29 pKa = 4.15MDD31 pKa = 3.14VTVRR35 pKa = 11.84PEE37 pKa = 3.46YY38 pKa = 9.26MAIVVTCGPVRR49 pKa = 11.84LWIKK53 pKa = 10.61APYY56 pKa = 9.49GSSLITAYY64 pKa = 10.59FPVSGYY70 pKa = 11.16AEE72 pKa = 4.16SCDD75 pKa = 3.22IEE77 pKa = 4.71HH78 pKa = 6.58EE79 pKa = 4.42NKK81 pKa = 10.35LGEE84 pKa = 4.68YY85 pKa = 8.71FTMIQSSDD93 pKa = 3.7MEE95 pKa = 4.1QLRR98 pKa = 11.84YY99 pKa = 10.61DD100 pKa = 3.07IMNWIRR106 pKa = 11.84VRR108 pKa = 11.84LVMSCLL114 pKa = 3.45
Molecular weight: 12.93 kDa
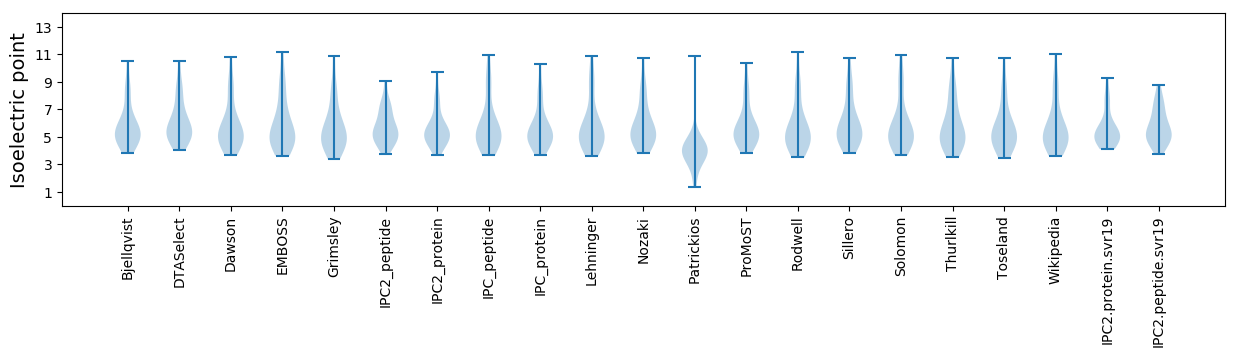
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G6Y069|A0A6G6Y069_9CAUD Uncharacterized protein OS=Bifidobacterium phage BadAargau2 OX=2713242 GN=BAAR0010003c01_00020 PE=4 SV=1
MM1 pKa = 7.42ARR3 pKa = 11.84SKK5 pKa = 9.96KK6 pKa = 4.7TTKK9 pKa = 10.01RR10 pKa = 11.84QRR12 pKa = 11.84QAAQRR17 pKa = 11.84KK18 pKa = 8.17RR19 pKa = 11.84NAQIRR24 pKa = 11.84SAQVARR30 pKa = 11.84NAARR34 pKa = 11.84NIGTINPTKK43 pKa = 10.73VSVGTLNKK51 pKa = 10.26LSRR54 pKa = 11.84PQLEE58 pKa = 3.69RR59 pKa = 11.84LAQNVGNEE67 pKa = 4.09VYY69 pKa = 10.08DD70 pKa = 4.03AKK72 pKa = 11.28SSVQKK77 pKa = 9.98YY78 pKa = 9.67LRR80 pKa = 11.84SKK82 pKa = 10.06PYY84 pKa = 8.6YY85 pKa = 8.63HH86 pKa = 7.34APPVSITKK94 pKa = 10.12RR95 pKa = 11.84EE96 pKa = 3.84RR97 pKa = 11.84EE98 pKa = 3.83FASRR102 pKa = 11.84KK103 pKa = 9.9VPTDD107 pKa = 3.28QQIAAAPPKK116 pKa = 9.7KK117 pKa = 10.34AKK119 pKa = 10.18LLRR122 pKa = 11.84QQKK125 pKa = 10.16RR126 pKa = 11.84KK127 pKa = 8.18ITAARR132 pKa = 11.84NKK134 pKa = 10.45LEE136 pKa = 3.62DD137 pKa = 3.23WKK139 pKa = 10.93RR140 pKa = 11.84YY141 pKa = 7.12NAQVMAGRR149 pKa = 11.84TARR152 pKa = 11.84DD153 pKa = 3.22EE154 pKa = 4.28RR155 pKa = 11.84ADD157 pKa = 3.66ALNGTGGKK165 pKa = 9.11EE166 pKa = 3.85SRR168 pKa = 11.84TRR170 pKa = 11.84DD171 pKa = 3.0AGKK174 pKa = 10.58RR175 pKa = 11.84STTDD179 pKa = 2.48IDD181 pKa = 3.53HH182 pKa = 7.57LDD184 pKa = 3.44RR185 pKa = 11.84YY186 pKa = 10.96RR187 pKa = 11.84NVLKK191 pKa = 10.81NPQIVSKK198 pKa = 11.11LSDD201 pKa = 3.47DD202 pKa = 4.03EE203 pKa = 4.04LRR205 pKa = 11.84KK206 pKa = 9.96EE207 pKa = 3.5IRR209 pKa = 11.84ARR211 pKa = 11.84TKK213 pKa = 10.49DD214 pKa = 3.45LKK216 pKa = 10.1NAKK219 pKa = 9.91NILEE223 pKa = 4.33NGVGRR228 pKa = 11.84ISEE231 pKa = 4.01KK232 pKa = 10.86DD233 pKa = 3.03NAYY236 pKa = 10.36VEE238 pKa = 4.48RR239 pKa = 11.84RR240 pKa = 11.84LKK242 pKa = 11.0AGFGQSALDD251 pKa = 3.47GWRR254 pKa = 11.84KK255 pKa = 9.63LSRR258 pKa = 11.84TEE260 pKa = 4.02RR261 pKa = 11.84YY262 pKa = 10.09SLVNQTSLLNRR273 pKa = 11.84IEE275 pKa = 4.6FYY277 pKa = 10.88SVYY280 pKa = 10.65NEE282 pKa = 3.69EE283 pKa = 4.03THH285 pKa = 7.47AYY287 pKa = 8.91EE288 pKa = 4.68PWDD291 pKa = 3.6HH292 pKa = 7.17DD293 pKa = 3.49RR294 pKa = 11.84KK295 pKa = 9.98EE296 pKa = 4.0QVKK299 pKa = 8.68QEE301 pKa = 4.13RR302 pKa = 11.84SDD304 pKa = 3.04IMSFIRR310 pKa = 11.84NARR313 pKa = 11.84NISNASLEE321 pKa = 4.16
MM1 pKa = 7.42ARR3 pKa = 11.84SKK5 pKa = 9.96KK6 pKa = 4.7TTKK9 pKa = 10.01RR10 pKa = 11.84QRR12 pKa = 11.84QAAQRR17 pKa = 11.84KK18 pKa = 8.17RR19 pKa = 11.84NAQIRR24 pKa = 11.84SAQVARR30 pKa = 11.84NAARR34 pKa = 11.84NIGTINPTKK43 pKa = 10.73VSVGTLNKK51 pKa = 10.26LSRR54 pKa = 11.84PQLEE58 pKa = 3.69RR59 pKa = 11.84LAQNVGNEE67 pKa = 4.09VYY69 pKa = 10.08DD70 pKa = 4.03AKK72 pKa = 11.28SSVQKK77 pKa = 9.98YY78 pKa = 9.67LRR80 pKa = 11.84SKK82 pKa = 10.06PYY84 pKa = 8.6YY85 pKa = 8.63HH86 pKa = 7.34APPVSITKK94 pKa = 10.12RR95 pKa = 11.84EE96 pKa = 3.84RR97 pKa = 11.84EE98 pKa = 3.83FASRR102 pKa = 11.84KK103 pKa = 9.9VPTDD107 pKa = 3.28QQIAAAPPKK116 pKa = 9.7KK117 pKa = 10.34AKK119 pKa = 10.18LLRR122 pKa = 11.84QQKK125 pKa = 10.16RR126 pKa = 11.84KK127 pKa = 8.18ITAARR132 pKa = 11.84NKK134 pKa = 10.45LEE136 pKa = 3.62DD137 pKa = 3.23WKK139 pKa = 10.93RR140 pKa = 11.84YY141 pKa = 7.12NAQVMAGRR149 pKa = 11.84TARR152 pKa = 11.84DD153 pKa = 3.22EE154 pKa = 4.28RR155 pKa = 11.84ADD157 pKa = 3.66ALNGTGGKK165 pKa = 9.11EE166 pKa = 3.85SRR168 pKa = 11.84TRR170 pKa = 11.84DD171 pKa = 3.0AGKK174 pKa = 10.58RR175 pKa = 11.84STTDD179 pKa = 2.48IDD181 pKa = 3.53HH182 pKa = 7.57LDD184 pKa = 3.44RR185 pKa = 11.84YY186 pKa = 10.96RR187 pKa = 11.84NVLKK191 pKa = 10.81NPQIVSKK198 pKa = 11.11LSDD201 pKa = 3.47DD202 pKa = 4.03EE203 pKa = 4.04LRR205 pKa = 11.84KK206 pKa = 9.96EE207 pKa = 3.5IRR209 pKa = 11.84ARR211 pKa = 11.84TKK213 pKa = 10.49DD214 pKa = 3.45LKK216 pKa = 10.1NAKK219 pKa = 9.91NILEE223 pKa = 4.33NGVGRR228 pKa = 11.84ISEE231 pKa = 4.01KK232 pKa = 10.86DD233 pKa = 3.03NAYY236 pKa = 10.36VEE238 pKa = 4.48RR239 pKa = 11.84RR240 pKa = 11.84LKK242 pKa = 11.0AGFGQSALDD251 pKa = 3.47GWRR254 pKa = 11.84KK255 pKa = 9.63LSRR258 pKa = 11.84TEE260 pKa = 4.02RR261 pKa = 11.84YY262 pKa = 10.09SLVNQTSLLNRR273 pKa = 11.84IEE275 pKa = 4.6FYY277 pKa = 10.88SVYY280 pKa = 10.65NEE282 pKa = 3.69EE283 pKa = 4.03THH285 pKa = 7.47AYY287 pKa = 8.91EE288 pKa = 4.68PWDD291 pKa = 3.6HH292 pKa = 7.17DD293 pKa = 3.49RR294 pKa = 11.84KK295 pKa = 9.98EE296 pKa = 4.0QVKK299 pKa = 8.68QEE301 pKa = 4.13RR302 pKa = 11.84SDD304 pKa = 3.04IMSFIRR310 pKa = 11.84NARR313 pKa = 11.84NISNASLEE321 pKa = 4.16
Molecular weight: 36.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5929 |
55 |
710 |
257.8 |
28.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.096 ± 0.587 | 0.978 ± 0.186 |
7.59 ± 0.503 | 4.992 ± 0.295 |
3.542 ± 0.339 | 6.831 ± 0.478 |
1.737 ± 0.213 | 5.768 ± 0.237 |
5.347 ± 0.472 | 6.612 ± 0.338 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.154 ± 0.286 | 6.409 ± 0.48 |
3.846 ± 0.326 | 4.284 ± 0.486 |
5.363 ± 0.55 | 6.814 ± 0.476 |
6.612 ± 0.451 | 5.65 ± 0.371 |
1.636 ± 0.145 | 4.739 ± 0.306 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
