
Pedobacter yulinensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Sphingobacteriia; Sphingobacteriales; Sphingobacteriaceae; Pedobacter
Average proteome isoelectric point is 7.08
Get precalculated fractions of proteins
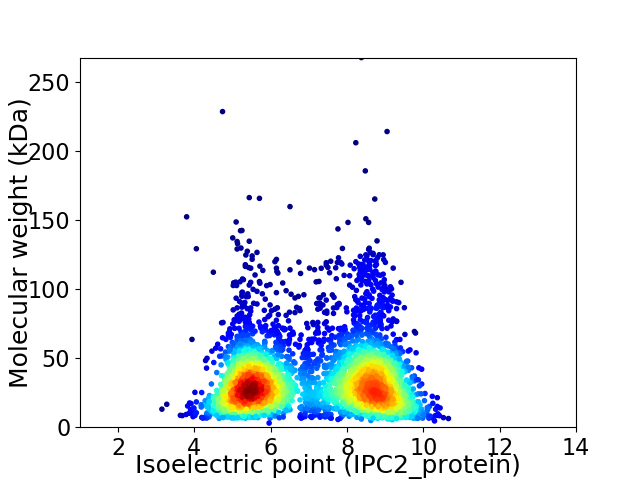
Virtual 2D-PAGE plot for 3772 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T3HND6|A0A2T3HND6_9SPHI Uncharacterized protein OS=Pedobacter yulinensis OX=2126353 GN=C7T94_03920 PE=4 SV=1
MM1 pKa = 6.72TQSDD5 pKa = 4.87LLALLQQGYY14 pKa = 8.99PGGAGSGSMDD24 pKa = 3.34ISTLQNPAYY33 pKa = 8.37TALFTQLGITSTTIPLNNSTGFAAEE58 pKa = 3.99GANIVFKK65 pKa = 11.4GNVNGSFFGLVNPQVTIQFSPDD87 pKa = 3.36SVNSGAFILLMHH99 pKa = 6.84FALPGAWTFSQSIPFFAGSIYY120 pKa = 9.82DD121 pKa = 3.85TLVLDD126 pKa = 4.13TSVAPALVMTSGPATDD142 pKa = 4.3PVNNAAATASGLYY155 pKa = 10.13FYY157 pKa = 11.26GLFSHH162 pKa = 6.3TQGPASFTLALSLLEE177 pKa = 4.6AFTSEE182 pKa = 4.12STVFGPVGLDD192 pKa = 2.97MSVPGQEE199 pKa = 4.53SINMQLALGSGTISNLFGQGSVSLTLSVTLFAGLLPGIDD238 pKa = 3.58QYY240 pKa = 12.02QKK242 pKa = 11.2GVLFTASLAFGQSTLNLGALLSGEE266 pKa = 4.05ADD268 pKa = 3.07GVLNLEE274 pKa = 4.36VTGTALAIDD283 pKa = 4.15EE284 pKa = 4.68SAINTYY290 pKa = 10.33FGSNNGIVSSLPGQFQTDD308 pKa = 3.41EE309 pKa = 4.21SLIAITAMNFGIGLTSHH326 pKa = 5.94NLEE329 pKa = 4.14YY330 pKa = 11.56AMMTISVLQGEE341 pKa = 5.06LWNVWMNGSTPVISIGNLAFTFSIFQPFGASPLYY375 pKa = 9.65EE376 pKa = 4.29FNFAGIFEE384 pKa = 5.1LGGVVPILIQAQLGNAEE401 pKa = 4.52DD402 pKa = 5.25LILSGQLQPGEE413 pKa = 4.3EE414 pKa = 4.31PGVAALIAAFYY425 pKa = 8.4PQQTTVPADD434 pKa = 3.52LVFEE438 pKa = 5.24KK439 pKa = 10.2IDD441 pKa = 3.69FQADD445 pKa = 3.01ISQGTFNADD454 pKa = 3.08LVIAGEE460 pKa = 4.26WEE462 pKa = 4.31FGFGDD467 pKa = 3.44NGSIAFEE474 pKa = 4.01EE475 pKa = 4.37LSMFFSYY482 pKa = 10.8DD483 pKa = 3.05SSNTPFIQAGVEE495 pKa = 4.37GIFRR499 pKa = 11.84IDD501 pKa = 3.15QTNQFDD507 pKa = 3.95IALTLSNTNFTFSGAWKK524 pKa = 8.9DD525 pKa = 3.21TGTPITYY532 pKa = 9.44IDD534 pKa = 3.25IVMALGLYY542 pKa = 9.15GLPQLPDD549 pKa = 3.14IDD551 pKa = 5.41LSLTGAKK558 pKa = 8.75LTFDD562 pKa = 3.73SQNLIFTFEE571 pKa = 4.57LDD573 pKa = 3.54SKK575 pKa = 11.57SFGDD579 pKa = 3.82ALIIAGKK586 pKa = 8.25STTGAWGFVFGLEE599 pKa = 4.07ATPKK603 pKa = 10.58GEE605 pKa = 5.11PINIDD610 pKa = 2.86ITNIPVVGNLVPAGDD625 pKa = 4.22DD626 pKa = 3.39TLSITNLRR634 pKa = 11.84FVGATNSLPAVTLSAAQQAIVGTQLSSGITLSIDD668 pKa = 3.6FAVGTLLSEE677 pKa = 4.29SFTVRR682 pKa = 11.84FALSDD687 pKa = 3.57GSQYY691 pKa = 9.15DD692 pKa = 3.96TPPPPPAQSTTLWQPPAPQQSGLPASAPAGSLATWVKK729 pKa = 8.53VQRR732 pKa = 11.84SFGPVQIDD740 pKa = 3.36SVGFQIVNGNSLALLVSGGVSLGGLSIGLTALEE773 pKa = 3.91AVIPISKK780 pKa = 9.72PFTPSFSLGGLEE792 pKa = 4.11VAYY795 pKa = 9.2TGTGFSVAGGLLTVPGKK812 pKa = 8.85TPAEE816 pKa = 3.93YY817 pKa = 10.59LGNLSVNAGPFGVTAFGMYY836 pKa = 8.81STVAGQPSLFAYY848 pKa = 10.21LAVSVPLGGPPFFFVTGLAGGFGYY872 pKa = 10.4NAQIQLPTIEE882 pKa = 4.88NVATYY887 pKa = 9.87PLIQAVTQTPPQSASQVQAEE907 pKa = 4.23LQQLVSTQNNQNWLAAGIGFSSFEE931 pKa = 4.05MLNSFALLTVSFGNTFAVALMGEE954 pKa = 4.54SSLSVPLSTKK964 pKa = 10.84DD965 pKa = 3.46EE966 pKa = 4.38IKK968 pKa = 10.74VEE970 pKa = 4.77PIAEE974 pKa = 4.05AQMEE978 pKa = 4.25ILIEE982 pKa = 3.86FLPSNGIFAVNAQLTPSSYY1001 pKa = 11.19VLSKK1005 pKa = 10.47NAKK1008 pKa = 7.79LTGGFAFDD1016 pKa = 4.06LWFSPSPYY1024 pKa = 10.53AGDD1027 pKa = 3.66FVITLGGYY1035 pKa = 8.53NPYY1038 pKa = 10.24YY1039 pKa = 10.53VPQKK1043 pKa = 10.21YY1044 pKa = 9.09YY1045 pKa = 10.13PAVPRR1050 pKa = 11.84LGISWQLSSLMSITGSAYY1068 pKa = 9.79FALTPSVIMAGGSLNAVYY1086 pKa = 10.01HH1087 pKa = 6.27AGNLRR1092 pKa = 11.84AWFTAEE1098 pKa = 3.61ADD1100 pKa = 3.45FLIRR1104 pKa = 11.84FKK1106 pKa = 10.58PFSFIVSISVTVGVSMKK1123 pKa = 10.75LNLLFTTTTITLSVGLGLQLWGLPFGGTARR1153 pKa = 11.84MDD1155 pKa = 3.61LSIISISINFGSSLSKK1171 pKa = 10.45PPEE1174 pKa = 4.05VNWDD1178 pKa = 3.04IFTEE1182 pKa = 4.41SFLPPPQSVPPAGQASPLAPQALDD1206 pKa = 3.34ATGVPVTTTDD1216 pKa = 3.96SILSLSASSGLLQSISTYY1234 pKa = 7.18TWQVDD1239 pKa = 3.37PEE1241 pKa = 4.6TFVMLLTLQIPATSAVVNTTDD1262 pKa = 2.95EE1263 pKa = 4.49HH1264 pKa = 6.4NNPVSTPVSGSWNTNLGVGPMGVGPGGLTAALTIQITAPAAGDD1307 pKa = 3.37EE1308 pKa = 4.56DD1309 pKa = 4.27VWVAQAVTGQVASGLWANTTNTMSTDD1335 pKa = 3.02GTLQNVLVGVSLVPVPPQPATTAAIDD1361 pKa = 4.22LSLLLYY1367 pKa = 10.82DD1368 pKa = 4.72PGNPVSWNWSSPQVVNTDD1386 pKa = 3.37PYY1388 pKa = 11.71GNDD1391 pKa = 2.84NPMAEE1396 pKa = 4.44MQSSLIDD1403 pKa = 3.29ATVAATRR1410 pKa = 11.84SSWINSLIAQGFALSNEE1427 pKa = 3.9VDD1429 pKa = 3.48VSDD1432 pKa = 5.35FSSNANNLLLSAPALRR1448 pKa = 11.84LAGEE1452 pKa = 4.35EE1453 pKa = 4.15KK1454 pKa = 10.91AEE1456 pKa = 4.21SEE1458 pKa = 4.36
MM1 pKa = 6.72TQSDD5 pKa = 4.87LLALLQQGYY14 pKa = 8.99PGGAGSGSMDD24 pKa = 3.34ISTLQNPAYY33 pKa = 8.37TALFTQLGITSTTIPLNNSTGFAAEE58 pKa = 3.99GANIVFKK65 pKa = 11.4GNVNGSFFGLVNPQVTIQFSPDD87 pKa = 3.36SVNSGAFILLMHH99 pKa = 6.84FALPGAWTFSQSIPFFAGSIYY120 pKa = 9.82DD121 pKa = 3.85TLVLDD126 pKa = 4.13TSVAPALVMTSGPATDD142 pKa = 4.3PVNNAAATASGLYY155 pKa = 10.13FYY157 pKa = 11.26GLFSHH162 pKa = 6.3TQGPASFTLALSLLEE177 pKa = 4.6AFTSEE182 pKa = 4.12STVFGPVGLDD192 pKa = 2.97MSVPGQEE199 pKa = 4.53SINMQLALGSGTISNLFGQGSVSLTLSVTLFAGLLPGIDD238 pKa = 3.58QYY240 pKa = 12.02QKK242 pKa = 11.2GVLFTASLAFGQSTLNLGALLSGEE266 pKa = 4.05ADD268 pKa = 3.07GVLNLEE274 pKa = 4.36VTGTALAIDD283 pKa = 4.15EE284 pKa = 4.68SAINTYY290 pKa = 10.33FGSNNGIVSSLPGQFQTDD308 pKa = 3.41EE309 pKa = 4.21SLIAITAMNFGIGLTSHH326 pKa = 5.94NLEE329 pKa = 4.14YY330 pKa = 11.56AMMTISVLQGEE341 pKa = 5.06LWNVWMNGSTPVISIGNLAFTFSIFQPFGASPLYY375 pKa = 9.65EE376 pKa = 4.29FNFAGIFEE384 pKa = 5.1LGGVVPILIQAQLGNAEE401 pKa = 4.52DD402 pKa = 5.25LILSGQLQPGEE413 pKa = 4.3EE414 pKa = 4.31PGVAALIAAFYY425 pKa = 8.4PQQTTVPADD434 pKa = 3.52LVFEE438 pKa = 5.24KK439 pKa = 10.2IDD441 pKa = 3.69FQADD445 pKa = 3.01ISQGTFNADD454 pKa = 3.08LVIAGEE460 pKa = 4.26WEE462 pKa = 4.31FGFGDD467 pKa = 3.44NGSIAFEE474 pKa = 4.01EE475 pKa = 4.37LSMFFSYY482 pKa = 10.8DD483 pKa = 3.05SSNTPFIQAGVEE495 pKa = 4.37GIFRR499 pKa = 11.84IDD501 pKa = 3.15QTNQFDD507 pKa = 3.95IALTLSNTNFTFSGAWKK524 pKa = 8.9DD525 pKa = 3.21TGTPITYY532 pKa = 9.44IDD534 pKa = 3.25IVMALGLYY542 pKa = 9.15GLPQLPDD549 pKa = 3.14IDD551 pKa = 5.41LSLTGAKK558 pKa = 8.75LTFDD562 pKa = 3.73SQNLIFTFEE571 pKa = 4.57LDD573 pKa = 3.54SKK575 pKa = 11.57SFGDD579 pKa = 3.82ALIIAGKK586 pKa = 8.25STTGAWGFVFGLEE599 pKa = 4.07ATPKK603 pKa = 10.58GEE605 pKa = 5.11PINIDD610 pKa = 2.86ITNIPVVGNLVPAGDD625 pKa = 4.22DD626 pKa = 3.39TLSITNLRR634 pKa = 11.84FVGATNSLPAVTLSAAQQAIVGTQLSSGITLSIDD668 pKa = 3.6FAVGTLLSEE677 pKa = 4.29SFTVRR682 pKa = 11.84FALSDD687 pKa = 3.57GSQYY691 pKa = 9.15DD692 pKa = 3.96TPPPPPAQSTTLWQPPAPQQSGLPASAPAGSLATWVKK729 pKa = 8.53VQRR732 pKa = 11.84SFGPVQIDD740 pKa = 3.36SVGFQIVNGNSLALLVSGGVSLGGLSIGLTALEE773 pKa = 3.91AVIPISKK780 pKa = 9.72PFTPSFSLGGLEE792 pKa = 4.11VAYY795 pKa = 9.2TGTGFSVAGGLLTVPGKK812 pKa = 8.85TPAEE816 pKa = 3.93YY817 pKa = 10.59LGNLSVNAGPFGVTAFGMYY836 pKa = 8.81STVAGQPSLFAYY848 pKa = 10.21LAVSVPLGGPPFFFVTGLAGGFGYY872 pKa = 10.4NAQIQLPTIEE882 pKa = 4.88NVATYY887 pKa = 9.87PLIQAVTQTPPQSASQVQAEE907 pKa = 4.23LQQLVSTQNNQNWLAAGIGFSSFEE931 pKa = 4.05MLNSFALLTVSFGNTFAVALMGEE954 pKa = 4.54SSLSVPLSTKK964 pKa = 10.84DD965 pKa = 3.46EE966 pKa = 4.38IKK968 pKa = 10.74VEE970 pKa = 4.77PIAEE974 pKa = 4.05AQMEE978 pKa = 4.25ILIEE982 pKa = 3.86FLPSNGIFAVNAQLTPSSYY1001 pKa = 11.19VLSKK1005 pKa = 10.47NAKK1008 pKa = 7.79LTGGFAFDD1016 pKa = 4.06LWFSPSPYY1024 pKa = 10.53AGDD1027 pKa = 3.66FVITLGGYY1035 pKa = 8.53NPYY1038 pKa = 10.24YY1039 pKa = 10.53VPQKK1043 pKa = 10.21YY1044 pKa = 9.09YY1045 pKa = 10.13PAVPRR1050 pKa = 11.84LGISWQLSSLMSITGSAYY1068 pKa = 9.79FALTPSVIMAGGSLNAVYY1086 pKa = 10.01HH1087 pKa = 6.27AGNLRR1092 pKa = 11.84AWFTAEE1098 pKa = 3.61ADD1100 pKa = 3.45FLIRR1104 pKa = 11.84FKK1106 pKa = 10.58PFSFIVSISVTVGVSMKK1123 pKa = 10.75LNLLFTTTTITLSVGLGLQLWGLPFGGTARR1153 pKa = 11.84MDD1155 pKa = 3.61LSIISISINFGSSLSKK1171 pKa = 10.45PPEE1174 pKa = 4.05VNWDD1178 pKa = 3.04IFTEE1182 pKa = 4.41SFLPPPQSVPPAGQASPLAPQALDD1206 pKa = 3.34ATGVPVTTTDD1216 pKa = 3.96SILSLSASSGLLQSISTYY1234 pKa = 7.18TWQVDD1239 pKa = 3.37PEE1241 pKa = 4.6TFVMLLTLQIPATSAVVNTTDD1262 pKa = 2.95EE1263 pKa = 4.49HH1264 pKa = 6.4NNPVSTPVSGSWNTNLGVGPMGVGPGGLTAALTIQITAPAAGDD1307 pKa = 3.37EE1308 pKa = 4.56DD1309 pKa = 4.27VWVAQAVTGQVASGLWANTTNTMSTDD1335 pKa = 3.02GTLQNVLVGVSLVPVPPQPATTAAIDD1361 pKa = 4.22LSLLLYY1367 pKa = 10.82DD1368 pKa = 4.72PGNPVSWNWSSPQVVNTDD1386 pKa = 3.37PYY1388 pKa = 11.71GNDD1391 pKa = 2.84NPMAEE1396 pKa = 4.44MQSSLIDD1403 pKa = 3.29ATVAATRR1410 pKa = 11.84SSWINSLIAQGFALSNEE1427 pKa = 3.9VDD1429 pKa = 3.48VSDD1432 pKa = 5.35FSSNANNLLLSAPALRR1448 pKa = 11.84LAGEE1452 pKa = 4.35EE1453 pKa = 4.15KK1454 pKa = 10.91AEE1456 pKa = 4.21SEE1458 pKa = 4.36
Molecular weight: 152.3 kDa
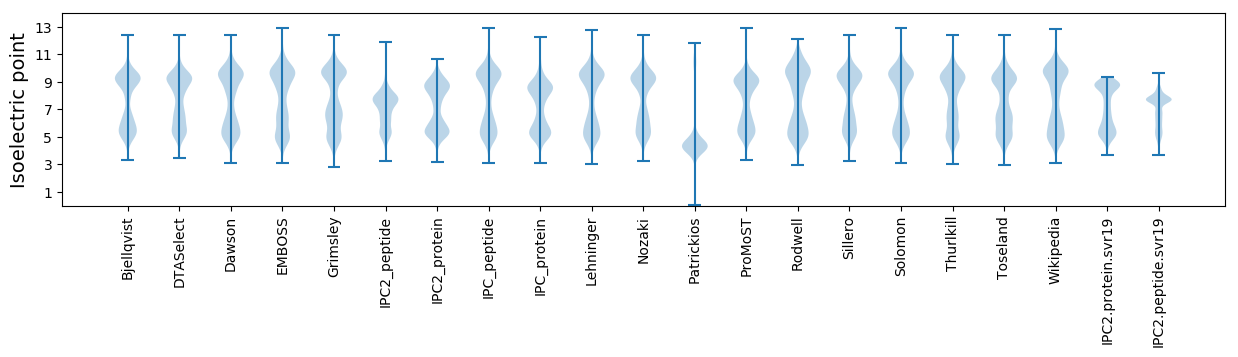
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T3HK36|A0A2T3HK36_9SPHI Uncharacterized protein OS=Pedobacter yulinensis OX=2126353 GN=C7T94_09135 PE=4 SV=1
MM1 pKa = 7.07KK2 pKa = 9.48TVCRR6 pKa = 11.84TRR8 pKa = 11.84CRR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 7.44SLRR16 pKa = 11.84NDD18 pKa = 2.56SSTRR22 pKa = 11.84GIGLRR27 pKa = 11.84SIAGRR32 pKa = 11.84VALLDD37 pKa = 3.67GSLTITCPEE46 pKa = 3.95KK47 pKa = 10.52GGTRR51 pKa = 11.84ILIRR55 pKa = 11.84FPLATVGQTGKK66 pKa = 10.37YY67 pKa = 8.79VPNASRR73 pKa = 11.84PAQLTDD79 pKa = 3.45LEE81 pKa = 4.61TDD83 pKa = 3.57GGQPAFYY90 pKa = 10.76GG91 pKa = 3.65
MM1 pKa = 7.07KK2 pKa = 9.48TVCRR6 pKa = 11.84TRR8 pKa = 11.84CRR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 7.44SLRR16 pKa = 11.84NDD18 pKa = 2.56SSTRR22 pKa = 11.84GIGLRR27 pKa = 11.84SIAGRR32 pKa = 11.84VALLDD37 pKa = 3.67GSLTITCPEE46 pKa = 3.95KK47 pKa = 10.52GGTRR51 pKa = 11.84ILIRR55 pKa = 11.84FPLATVGQTGKK66 pKa = 10.37YY67 pKa = 8.79VPNASRR73 pKa = 11.84PAQLTDD79 pKa = 3.45LEE81 pKa = 4.61TDD83 pKa = 3.57GGQPAFYY90 pKa = 10.76GG91 pKa = 3.65
Molecular weight: 9.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1296689 |
25 |
2386 |
343.8 |
38.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.866 ± 0.041 | 0.794 ± 0.012 |
5.167 ± 0.026 | 5.606 ± 0.043 |
4.873 ± 0.027 | 7.403 ± 0.035 |
1.899 ± 0.023 | 6.061 ± 0.035 |
5.747 ± 0.035 | 9.91 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.275 ± 0.017 | 4.826 ± 0.043 |
4.159 ± 0.024 | 3.92 ± 0.024 |
5.326 ± 0.031 | 6.161 ± 0.03 |
5.507 ± 0.038 | 6.484 ± 0.031 |
1.214 ± 0.018 | 3.801 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
