
Marinobacter vinifirmus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Marinobacter
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
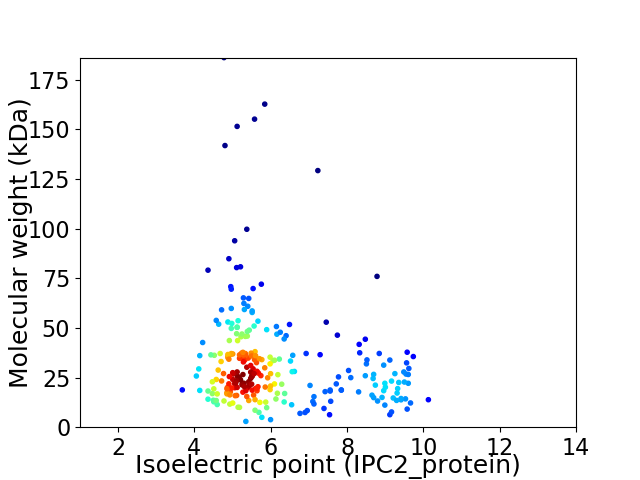
Virtual 2D-PAGE plot for 263 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
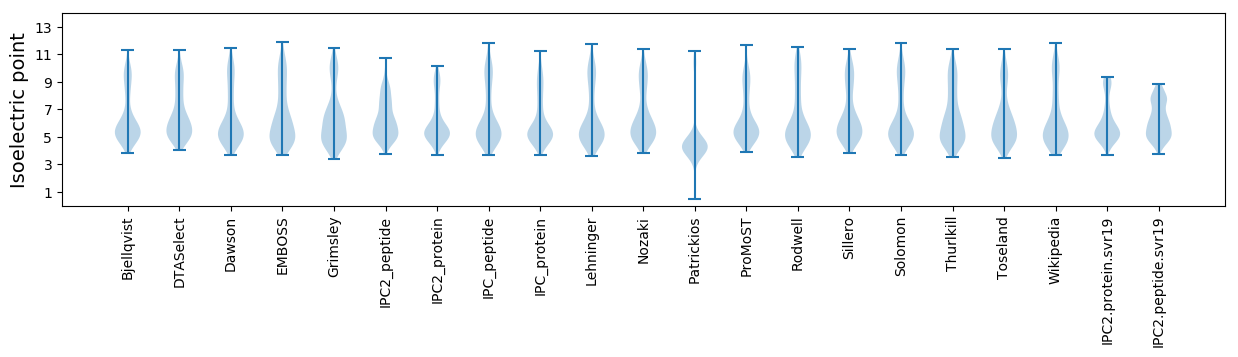
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T4CRA7|A0A2T4CRA7_9ALTE Polymerase (Fragment) OS=Marinobacter vinifirmus OX=355591 GN=C9985_01025 PE=4 SV=1
MM1 pKa = 7.49TIKK4 pKa = 10.68KK5 pKa = 9.28NVLIASVATAGIAMAGAASAQDD27 pKa = 3.74MYY29 pKa = 11.44KK30 pKa = 10.43SGVGGLYY37 pKa = 10.58AGGNYY42 pKa = 9.16TFMNADD48 pKa = 3.26FDD50 pKa = 4.68GVGDD54 pKa = 4.22ADD56 pKa = 3.76VGTLSGKK63 pKa = 9.25VGVMATEE70 pKa = 4.07YY71 pKa = 10.64FGLEE75 pKa = 3.49ARR77 pKa = 11.84AGFGVDD83 pKa = 3.47DD84 pKa = 3.93DD85 pKa = 4.4TVRR88 pKa = 11.84GVDD91 pKa = 3.49VEE93 pKa = 4.24LDD95 pKa = 3.6NFFGGYY101 pKa = 8.27ATFNMVNEE109 pKa = 4.45SPITPYY115 pKa = 10.93GVLGFTRR122 pKa = 11.84IEE124 pKa = 3.94AQAGNVEE131 pKa = 4.9DD132 pKa = 5.17DD133 pKa = 5.24DD134 pKa = 6.03SDD136 pKa = 3.72FSYY139 pKa = 11.1GAGVNFAFAPNLSANLEE156 pKa = 4.0YY157 pKa = 10.04MRR159 pKa = 11.84YY160 pKa = 9.91YY161 pKa = 10.81DD162 pKa = 5.2DD163 pKa = 5.33NDD165 pKa = 3.49VTVDD169 pKa = 3.44GLGIGMQVNFF179 pKa = 4.47
MM1 pKa = 7.49TIKK4 pKa = 10.68KK5 pKa = 9.28NVLIASVATAGIAMAGAASAQDD27 pKa = 3.74MYY29 pKa = 11.44KK30 pKa = 10.43SGVGGLYY37 pKa = 10.58AGGNYY42 pKa = 9.16TFMNADD48 pKa = 3.26FDD50 pKa = 4.68GVGDD54 pKa = 4.22ADD56 pKa = 3.76VGTLSGKK63 pKa = 9.25VGVMATEE70 pKa = 4.07YY71 pKa = 10.64FGLEE75 pKa = 3.49ARR77 pKa = 11.84AGFGVDD83 pKa = 3.47DD84 pKa = 3.93DD85 pKa = 4.4TVRR88 pKa = 11.84GVDD91 pKa = 3.49VEE93 pKa = 4.24LDD95 pKa = 3.6NFFGGYY101 pKa = 8.27ATFNMVNEE109 pKa = 4.45SPITPYY115 pKa = 10.93GVLGFTRR122 pKa = 11.84IEE124 pKa = 3.94AQAGNVEE131 pKa = 4.9DD132 pKa = 5.17DD133 pKa = 5.24DD134 pKa = 6.03SDD136 pKa = 3.72FSYY139 pKa = 11.1GAGVNFAFAPNLSANLEE156 pKa = 4.0YY157 pKa = 10.04MRR159 pKa = 11.84YY160 pKa = 9.91YY161 pKa = 10.81DD162 pKa = 5.2DD163 pKa = 5.33NDD165 pKa = 3.49VTVDD169 pKa = 3.44GLGIGMQVNFF179 pKa = 4.47
Molecular weight: 18.79 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T4CRF6|A0A2T4CRF6_9ALTE Tripartite tricarboxylate transporter TctB family protein OS=Marinobacter vinifirmus OX=355591 GN=C9985_00865 PE=4 SV=1
MM1 pKa = 7.5IMHH4 pKa = 7.85LFRR7 pKa = 11.84VSPDD11 pKa = 3.36DD12 pKa = 3.92DD13 pKa = 3.99ALRR16 pKa = 11.84GALLMTGAAILFAAMAVLIRR36 pKa = 11.84YY37 pKa = 6.59ITRR40 pKa = 11.84EE41 pKa = 3.73VHH43 pKa = 5.55PFEE46 pKa = 5.03AAFFRR51 pKa = 11.84NLFGLIPMIPWMLRR65 pKa = 11.84DD66 pKa = 3.56RR67 pKa = 11.84LAGLRR72 pKa = 11.84TLKK75 pKa = 10.77LKK77 pKa = 10.63LHH79 pKa = 7.21FIRR82 pKa = 11.84AVIGLAGMLCLFSALAIAPAAQVIAINFTVPILTTILAALILHH125 pKa = 5.3EE126 pKa = 4.4TVRR129 pKa = 11.84ARR131 pKa = 11.84RR132 pKa = 11.84WTAVALGFIGAMIIVRR148 pKa = 11.84PIGQTLEE155 pKa = 3.96TGAILALLATLFTAFAVTTVKK176 pKa = 9.92MLSRR180 pKa = 11.84SEE182 pKa = 4.06SANAIVTWMGMIMTPLSLIPALFFWQNPTWEE213 pKa = 4.15QLGVLLAIAVLATAGQQLFVRR234 pKa = 11.84ANRR237 pKa = 11.84TADD240 pKa = 3.11QSYY243 pKa = 10.79VMAFDD248 pKa = 4.27FLRR251 pKa = 11.84LPFVAALAFIMFGEE265 pKa = 4.71TVDD268 pKa = 3.87FWTWAGASLIIGSTLYY284 pKa = 9.74IARR287 pKa = 11.84RR288 pKa = 11.84EE289 pKa = 3.91AVLARR294 pKa = 11.84KK295 pKa = 8.29EE296 pKa = 3.81KK297 pKa = 10.14RR298 pKa = 11.84RR299 pKa = 11.84RR300 pKa = 11.84RR301 pKa = 11.84MPTTAAADD309 pKa = 3.69SQAIPVTKK317 pKa = 10.21AKK319 pKa = 9.81TINPPEE325 pKa = 4.25YY326 pKa = 10.15PEE328 pKa = 4.44GTPEE332 pKa = 4.29SNTGNKK338 pKa = 8.12TGRR341 pKa = 11.84IDD343 pKa = 3.13AA344 pKa = 4.83
MM1 pKa = 7.5IMHH4 pKa = 7.85LFRR7 pKa = 11.84VSPDD11 pKa = 3.36DD12 pKa = 3.92DD13 pKa = 3.99ALRR16 pKa = 11.84GALLMTGAAILFAAMAVLIRR36 pKa = 11.84YY37 pKa = 6.59ITRR40 pKa = 11.84EE41 pKa = 3.73VHH43 pKa = 5.55PFEE46 pKa = 5.03AAFFRR51 pKa = 11.84NLFGLIPMIPWMLRR65 pKa = 11.84DD66 pKa = 3.56RR67 pKa = 11.84LAGLRR72 pKa = 11.84TLKK75 pKa = 10.77LKK77 pKa = 10.63LHH79 pKa = 7.21FIRR82 pKa = 11.84AVIGLAGMLCLFSALAIAPAAQVIAINFTVPILTTILAALILHH125 pKa = 5.3EE126 pKa = 4.4TVRR129 pKa = 11.84ARR131 pKa = 11.84RR132 pKa = 11.84WTAVALGFIGAMIIVRR148 pKa = 11.84PIGQTLEE155 pKa = 3.96TGAILALLATLFTAFAVTTVKK176 pKa = 9.92MLSRR180 pKa = 11.84SEE182 pKa = 4.06SANAIVTWMGMIMTPLSLIPALFFWQNPTWEE213 pKa = 4.15QLGVLLAIAVLATAGQQLFVRR234 pKa = 11.84ANRR237 pKa = 11.84TADD240 pKa = 3.11QSYY243 pKa = 10.79VMAFDD248 pKa = 4.27FLRR251 pKa = 11.84LPFVAALAFIMFGEE265 pKa = 4.71TVDD268 pKa = 3.87FWTWAGASLIIGSTLYY284 pKa = 9.74IARR287 pKa = 11.84RR288 pKa = 11.84EE289 pKa = 3.91AVLARR294 pKa = 11.84KK295 pKa = 8.29EE296 pKa = 3.81KK297 pKa = 10.14RR298 pKa = 11.84RR299 pKa = 11.84RR300 pKa = 11.84RR301 pKa = 11.84MPTTAAADD309 pKa = 3.69SQAIPVTKK317 pKa = 10.21AKK319 pKa = 9.81TINPPEE325 pKa = 4.25YY326 pKa = 10.15PEE328 pKa = 4.44GTPEE332 pKa = 4.29SNTGNKK338 pKa = 8.12TGRR341 pKa = 11.84IDD343 pKa = 3.13AA344 pKa = 4.83
Molecular weight: 37.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
78148 |
25 |
1690 |
297.1 |
32.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.298 ± 0.174 | 0.909 ± 0.049 |
5.908 ± 0.16 | 6.023 ± 0.173 |
4.137 ± 0.13 | 8.118 ± 0.168 |
2.07 ± 0.075 | 6.063 ± 0.125 |
3.751 ± 0.125 | 10.112 ± 0.17 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.749 ± 0.077 | 3.291 ± 0.075 |
4.714 ± 0.1 | 3.404 ± 0.095 |
6.222 ± 0.138 | 5.675 ± 0.111 |
5.266 ± 0.084 | 7.449 ± 0.14 |
1.319 ± 0.068 | 2.523 ± 0.074 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
