
Flavilitoribacter nigricans DSM 23189 = NBRC 102662
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Saprospiria; Saprospirales; Lewinellaceae; Flavilitoribacter; Flavilitoribacter nigricans
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
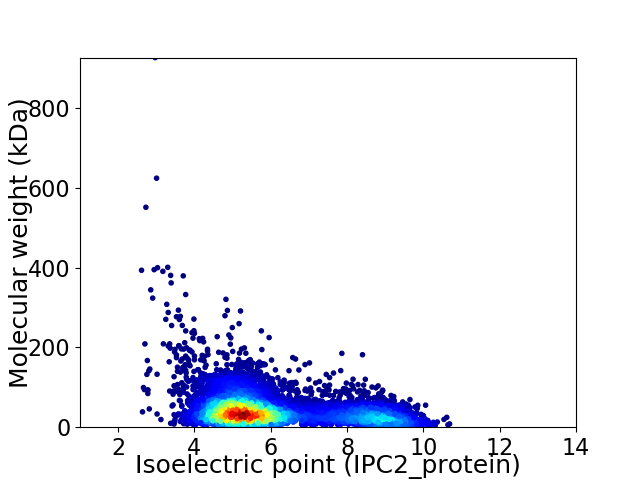
Virtual 2D-PAGE plot for 8179 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D0N4K6|A0A2D0N4K6_9BACT ATP phosphoribosyltransferase OS=Flavilitoribacter nigricans DSM 23189 = NBRC 102662 OX=1122177 GN=hisG PE=3 SV=1
MM1 pKa = 7.64TILEE5 pKa = 4.53KK6 pKa = 11.08YY7 pKa = 9.75NYY9 pKa = 8.96PIKK12 pKa = 9.96TLFAALILVGGFSCTRR28 pKa = 11.84DD29 pKa = 3.47LDD31 pKa = 4.07EE32 pKa = 6.75LEE34 pKa = 4.96LATFPTTAEE43 pKa = 3.86VFIDD47 pKa = 4.31GFSSGLNYY55 pKa = 10.6AAFGGSKK62 pKa = 6.79TTAFNVVDD70 pKa = 3.95DD71 pKa = 4.42EE72 pKa = 5.16GYY74 pKa = 10.84NNSAVMRR81 pKa = 11.84FDD83 pKa = 5.1IPDD86 pKa = 3.88ADD88 pKa = 4.05DD89 pKa = 4.4PGGGFAAGIFFTDD102 pKa = 3.28VARR105 pKa = 11.84DD106 pKa = 3.41LSGYY110 pKa = 10.31NVLTFWAKK118 pKa = 10.2ASEE121 pKa = 4.34SVDD124 pKa = 3.2VDD126 pKa = 3.34EE127 pKa = 6.15LGFGFTFQSEE137 pKa = 4.9KK138 pKa = 10.09YY139 pKa = 9.08RR140 pKa = 11.84SAIKK144 pKa = 9.71EE145 pKa = 4.06IPVTTNWKK153 pKa = 9.93KK154 pKa = 10.8YY155 pKa = 8.79YY156 pKa = 10.11IPIADD161 pKa = 3.48ASKK164 pKa = 8.89LTEE167 pKa = 4.16EE168 pKa = 4.47NGMFYY173 pKa = 10.76YY174 pKa = 10.54VDD176 pKa = 3.99SPDD179 pKa = 4.06DD180 pKa = 4.02GEE182 pKa = 6.28GYY184 pKa = 8.29TFWIDD189 pKa = 3.32EE190 pKa = 4.44VKK192 pKa = 10.77FEE194 pKa = 4.48NLSTLARR201 pKa = 11.84PGVRR205 pKa = 11.84ILAGQDD211 pKa = 3.19QSITAEE217 pKa = 4.11TGNQLNIGEE226 pKa = 4.24ISYY229 pKa = 8.16TANLPTGIDD238 pKa = 3.33QSVDD242 pKa = 2.44ISAAYY247 pKa = 8.47LTFTSSNSGVATVDD261 pKa = 3.1EE262 pKa = 4.58TGKK265 pKa = 8.65VTILDD270 pKa = 3.84AGNTVITATLGDD282 pKa = 4.2LEE284 pKa = 4.55ATGSLTIEE292 pKa = 4.27SSGDD296 pKa = 3.35PLLPATAAPVPTVGQDD312 pKa = 3.37SVISLFSNAYY322 pKa = 10.24DD323 pKa = 4.22DD324 pKa = 5.17VPVDD328 pKa = 3.06TWNPFWQFSTAEE340 pKa = 4.0VSDD343 pKa = 3.82VKK345 pKa = 10.78IGEE348 pKa = 4.78DD349 pKa = 3.85DD350 pKa = 3.77LKK352 pKa = 11.04RR353 pKa = 11.84YY354 pKa = 10.35NNLNFVGILTEE365 pKa = 4.12SAPIDD370 pKa = 3.87ASEE373 pKa = 4.1MTHH376 pKa = 5.27FHH378 pKa = 6.31IDD380 pKa = 2.35IWTPDD385 pKa = 3.47PTAPPATFKK394 pKa = 11.21VLLVDD399 pKa = 5.83FGADD403 pKa = 3.35GNFDD407 pKa = 3.79GGDD410 pKa = 3.76DD411 pKa = 3.94SSHH414 pKa = 6.33EE415 pKa = 4.17LTFTSPTLTTGSWVSLDD432 pKa = 3.4IPLSDD437 pKa = 3.64FTGLANRR444 pKa = 11.84NHH446 pKa = 6.87IAQLVLSGDD455 pKa = 3.92LPNLFVDD462 pKa = 4.19NVYY465 pKa = 10.2FYY467 pKa = 10.7QAGVVPVAEE476 pKa = 4.36PAEE479 pKa = 4.29AAPEE483 pKa = 3.94PTAAASDD490 pKa = 4.0VLSVFSDD497 pKa = 3.86AYY499 pKa = 11.43SNIEE503 pKa = 3.96GTDD506 pKa = 3.68LNPNWGQATVVSQVDD521 pKa = 3.21IAGNNTLLYY530 pKa = 10.75SGLNYY535 pKa = 10.27QGIQLGSNQDD545 pKa = 3.23VSGMEE550 pKa = 4.37FLHH553 pKa = 7.25IDD555 pKa = 3.14YY556 pKa = 7.68WTANSSGLNTFLISRR571 pKa = 11.84GPVEE575 pKa = 3.77TAYY578 pKa = 10.65ALPVPTTGWASVDD591 pKa = 3.12IPLSDD596 pKa = 4.53FAPVDD601 pKa = 3.88LTDD604 pKa = 4.37VIQLKK609 pKa = 10.55FDD611 pKa = 3.94GNGNIYY617 pKa = 10.33LDD619 pKa = 3.79NIYY622 pKa = 10.24FYY624 pKa = 9.67KK625 pKa = 10.32TGGGGMEE632 pKa = 4.34PAVSAPQPTEE642 pKa = 3.52AASEE646 pKa = 4.44VIALFSDD653 pKa = 4.36AYY655 pKa = 10.75ADD657 pKa = 3.63VVVDD661 pKa = 3.37TWRR664 pKa = 11.84TDD666 pKa = 2.31WSAAMLEE673 pKa = 4.35DD674 pKa = 4.04VSVAGNAVKK683 pKa = 10.29KK684 pKa = 10.54YY685 pKa = 10.73SNLDD689 pKa = 3.27FVGIEE694 pKa = 4.24TVANTVDD701 pKa = 3.51ASGMTHH707 pKa = 6.17FRR709 pKa = 11.84MDD711 pKa = 2.66VWSPDD716 pKa = 3.14FTFFGIKK723 pKa = 10.18LVDD726 pKa = 4.28FGTDD730 pKa = 3.15GAFGGGDD737 pKa = 3.43DD738 pKa = 5.13VEE740 pKa = 4.36HH741 pKa = 6.55QINFEE746 pKa = 4.24APAQGQWISYY756 pKa = 8.29DD757 pKa = 3.22IPLSDD762 pKa = 4.22FAGLTTRR769 pKa = 11.84SNIAQYY775 pKa = 10.95ILVGQPTGTTTVYY788 pKa = 10.5VDD790 pKa = 3.17NVYY793 pKa = 10.43FYY795 pKa = 11.43NSEE798 pKa = 3.92GGNNATEE805 pKa = 4.62PGLAAPTPTEE815 pKa = 4.22DD816 pKa = 3.51ASDD819 pKa = 4.04VISLFSEE826 pKa = 5.09AYY828 pKa = 10.47SNVAVDD834 pKa = 3.19TWRR837 pKa = 11.84TDD839 pKa = 2.3WSAAMLEE846 pKa = 4.35DD847 pKa = 4.04VSVAGNAVKK856 pKa = 10.29KK857 pKa = 10.54YY858 pKa = 10.73SNLDD862 pKa = 3.27FVGIEE867 pKa = 4.19TVANPIDD874 pKa = 4.17ASGMTYY880 pKa = 9.57FHH882 pKa = 7.07IDD884 pKa = 2.69VWSSDD889 pKa = 3.44FTFFGIKK896 pKa = 10.18LVDD899 pKa = 4.74FGADD903 pKa = 3.24GAFGGGDD910 pKa = 3.43DD911 pKa = 5.13VEE913 pKa = 4.36HH914 pKa = 6.55QINFEE919 pKa = 4.24APAQGQWISYY929 pKa = 8.36DD930 pKa = 3.17IPLDD934 pKa = 3.9DD935 pKa = 5.23FVGLTTRR942 pKa = 11.84SNMAQYY948 pKa = 10.75ILVGQPIGATTVYY961 pKa = 10.07VDD963 pKa = 2.98NVYY966 pKa = 10.29FHH968 pKa = 7.41KK969 pKa = 10.95
MM1 pKa = 7.64TILEE5 pKa = 4.53KK6 pKa = 11.08YY7 pKa = 9.75NYY9 pKa = 8.96PIKK12 pKa = 9.96TLFAALILVGGFSCTRR28 pKa = 11.84DD29 pKa = 3.47LDD31 pKa = 4.07EE32 pKa = 6.75LEE34 pKa = 4.96LATFPTTAEE43 pKa = 3.86VFIDD47 pKa = 4.31GFSSGLNYY55 pKa = 10.6AAFGGSKK62 pKa = 6.79TTAFNVVDD70 pKa = 3.95DD71 pKa = 4.42EE72 pKa = 5.16GYY74 pKa = 10.84NNSAVMRR81 pKa = 11.84FDD83 pKa = 5.1IPDD86 pKa = 3.88ADD88 pKa = 4.05DD89 pKa = 4.4PGGGFAAGIFFTDD102 pKa = 3.28VARR105 pKa = 11.84DD106 pKa = 3.41LSGYY110 pKa = 10.31NVLTFWAKK118 pKa = 10.2ASEE121 pKa = 4.34SVDD124 pKa = 3.2VDD126 pKa = 3.34EE127 pKa = 6.15LGFGFTFQSEE137 pKa = 4.9KK138 pKa = 10.09YY139 pKa = 9.08RR140 pKa = 11.84SAIKK144 pKa = 9.71EE145 pKa = 4.06IPVTTNWKK153 pKa = 9.93KK154 pKa = 10.8YY155 pKa = 8.79YY156 pKa = 10.11IPIADD161 pKa = 3.48ASKK164 pKa = 8.89LTEE167 pKa = 4.16EE168 pKa = 4.47NGMFYY173 pKa = 10.76YY174 pKa = 10.54VDD176 pKa = 3.99SPDD179 pKa = 4.06DD180 pKa = 4.02GEE182 pKa = 6.28GYY184 pKa = 8.29TFWIDD189 pKa = 3.32EE190 pKa = 4.44VKK192 pKa = 10.77FEE194 pKa = 4.48NLSTLARR201 pKa = 11.84PGVRR205 pKa = 11.84ILAGQDD211 pKa = 3.19QSITAEE217 pKa = 4.11TGNQLNIGEE226 pKa = 4.24ISYY229 pKa = 8.16TANLPTGIDD238 pKa = 3.33QSVDD242 pKa = 2.44ISAAYY247 pKa = 8.47LTFTSSNSGVATVDD261 pKa = 3.1EE262 pKa = 4.58TGKK265 pKa = 8.65VTILDD270 pKa = 3.84AGNTVITATLGDD282 pKa = 4.2LEE284 pKa = 4.55ATGSLTIEE292 pKa = 4.27SSGDD296 pKa = 3.35PLLPATAAPVPTVGQDD312 pKa = 3.37SVISLFSNAYY322 pKa = 10.24DD323 pKa = 4.22DD324 pKa = 5.17VPVDD328 pKa = 3.06TWNPFWQFSTAEE340 pKa = 4.0VSDD343 pKa = 3.82VKK345 pKa = 10.78IGEE348 pKa = 4.78DD349 pKa = 3.85DD350 pKa = 3.77LKK352 pKa = 11.04RR353 pKa = 11.84YY354 pKa = 10.35NNLNFVGILTEE365 pKa = 4.12SAPIDD370 pKa = 3.87ASEE373 pKa = 4.1MTHH376 pKa = 5.27FHH378 pKa = 6.31IDD380 pKa = 2.35IWTPDD385 pKa = 3.47PTAPPATFKK394 pKa = 11.21VLLVDD399 pKa = 5.83FGADD403 pKa = 3.35GNFDD407 pKa = 3.79GGDD410 pKa = 3.76DD411 pKa = 3.94SSHH414 pKa = 6.33EE415 pKa = 4.17LTFTSPTLTTGSWVSLDD432 pKa = 3.4IPLSDD437 pKa = 3.64FTGLANRR444 pKa = 11.84NHH446 pKa = 6.87IAQLVLSGDD455 pKa = 3.92LPNLFVDD462 pKa = 4.19NVYY465 pKa = 10.2FYY467 pKa = 10.7QAGVVPVAEE476 pKa = 4.36PAEE479 pKa = 4.29AAPEE483 pKa = 3.94PTAAASDD490 pKa = 4.0VLSVFSDD497 pKa = 3.86AYY499 pKa = 11.43SNIEE503 pKa = 3.96GTDD506 pKa = 3.68LNPNWGQATVVSQVDD521 pKa = 3.21IAGNNTLLYY530 pKa = 10.75SGLNYY535 pKa = 10.27QGIQLGSNQDD545 pKa = 3.23VSGMEE550 pKa = 4.37FLHH553 pKa = 7.25IDD555 pKa = 3.14YY556 pKa = 7.68WTANSSGLNTFLISRR571 pKa = 11.84GPVEE575 pKa = 3.77TAYY578 pKa = 10.65ALPVPTTGWASVDD591 pKa = 3.12IPLSDD596 pKa = 4.53FAPVDD601 pKa = 3.88LTDD604 pKa = 4.37VIQLKK609 pKa = 10.55FDD611 pKa = 3.94GNGNIYY617 pKa = 10.33LDD619 pKa = 3.79NIYY622 pKa = 10.24FYY624 pKa = 9.67KK625 pKa = 10.32TGGGGMEE632 pKa = 4.34PAVSAPQPTEE642 pKa = 3.52AASEE646 pKa = 4.44VIALFSDD653 pKa = 4.36AYY655 pKa = 10.75ADD657 pKa = 3.63VVVDD661 pKa = 3.37TWRR664 pKa = 11.84TDD666 pKa = 2.31WSAAMLEE673 pKa = 4.35DD674 pKa = 4.04VSVAGNAVKK683 pKa = 10.29KK684 pKa = 10.54YY685 pKa = 10.73SNLDD689 pKa = 3.27FVGIEE694 pKa = 4.24TVANTVDD701 pKa = 3.51ASGMTHH707 pKa = 6.17FRR709 pKa = 11.84MDD711 pKa = 2.66VWSPDD716 pKa = 3.14FTFFGIKK723 pKa = 10.18LVDD726 pKa = 4.28FGTDD730 pKa = 3.15GAFGGGDD737 pKa = 3.43DD738 pKa = 5.13VEE740 pKa = 4.36HH741 pKa = 6.55QINFEE746 pKa = 4.24APAQGQWISYY756 pKa = 8.29DD757 pKa = 3.22IPLSDD762 pKa = 4.22FAGLTTRR769 pKa = 11.84SNIAQYY775 pKa = 10.95ILVGQPTGTTTVYY788 pKa = 10.5VDD790 pKa = 3.17NVYY793 pKa = 10.43FYY795 pKa = 11.43NSEE798 pKa = 3.92GGNNATEE805 pKa = 4.62PGLAAPTPTEE815 pKa = 4.22DD816 pKa = 3.51ASDD819 pKa = 4.04VISLFSEE826 pKa = 5.09AYY828 pKa = 10.47SNVAVDD834 pKa = 3.19TWRR837 pKa = 11.84TDD839 pKa = 2.3WSAAMLEE846 pKa = 4.35DD847 pKa = 4.04VSVAGNAVKK856 pKa = 10.29KK857 pKa = 10.54YY858 pKa = 10.73SNLDD862 pKa = 3.27FVGIEE867 pKa = 4.19TVANPIDD874 pKa = 4.17ASGMTYY880 pKa = 9.57FHH882 pKa = 7.07IDD884 pKa = 2.69VWSSDD889 pKa = 3.44FTFFGIKK896 pKa = 10.18LVDD899 pKa = 4.74FGADD903 pKa = 3.24GAFGGGDD910 pKa = 3.43DD911 pKa = 5.13VEE913 pKa = 4.36HH914 pKa = 6.55QINFEE919 pKa = 4.24APAQGQWISYY929 pKa = 8.36DD930 pKa = 3.17IPLDD934 pKa = 3.9DD935 pKa = 5.23FVGLTTRR942 pKa = 11.84SNMAQYY948 pKa = 10.75ILVGQPIGATTVYY961 pKa = 10.07VDD963 pKa = 2.98NVYY966 pKa = 10.29FHH968 pKa = 7.41KK969 pKa = 10.95
Molecular weight: 104.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D0NFV0|A0A2D0NFV0_9BACT Uncharacterized protein OS=Flavilitoribacter nigricans DSM 23189 = NBRC 102662 OX=1122177 GN=CRP01_06400 PE=4 SV=1
MM1 pKa = 7.52HH2 pKa = 7.31SGFGAEE8 pKa = 4.07VLLIARR14 pKa = 11.84MPKK17 pKa = 10.13PMLRR21 pKa = 11.84QCSCSSAWFRR31 pKa = 11.84LPKK34 pKa = 10.34AEE36 pKa = 4.39PRR38 pKa = 11.84GLLIRR43 pKa = 11.84WINIQAPYY51 pKa = 10.56ASEE54 pKa = 4.51LLGQATIRR62 pKa = 11.84KK63 pKa = 7.92NHH65 pKa = 6.05FSALQILPFLFF76 pKa = 4.38
MM1 pKa = 7.52HH2 pKa = 7.31SGFGAEE8 pKa = 4.07VLLIARR14 pKa = 11.84MPKK17 pKa = 10.13PMLRR21 pKa = 11.84QCSCSSAWFRR31 pKa = 11.84LPKK34 pKa = 10.34AEE36 pKa = 4.39PRR38 pKa = 11.84GLLIRR43 pKa = 11.84WINIQAPYY51 pKa = 10.56ASEE54 pKa = 4.51LLGQATIRR62 pKa = 11.84KK63 pKa = 7.92NHH65 pKa = 6.05FSALQILPFLFF76 pKa = 4.38
Molecular weight: 8.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3270642 |
38 |
8758 |
399.9 |
44.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.554 ± 0.025 | 0.919 ± 0.022 |
6.168 ± 0.038 | 6.573 ± 0.026 |
4.715 ± 0.021 | 7.336 ± 0.029 |
1.919 ± 0.016 | 6.331 ± 0.021 |
4.659 ± 0.039 | 9.888 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.123 ± 0.015 | 4.603 ± 0.022 |
4.499 ± 0.023 | 4.163 ± 0.022 |
5.267 ± 0.036 | 6.046 ± 0.025 |
5.706 ± 0.063 | 6.166 ± 0.03 |
1.451 ± 0.011 | 3.914 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
