
Prochlorococcus phage P-SSP7
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Tiamatvirus
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
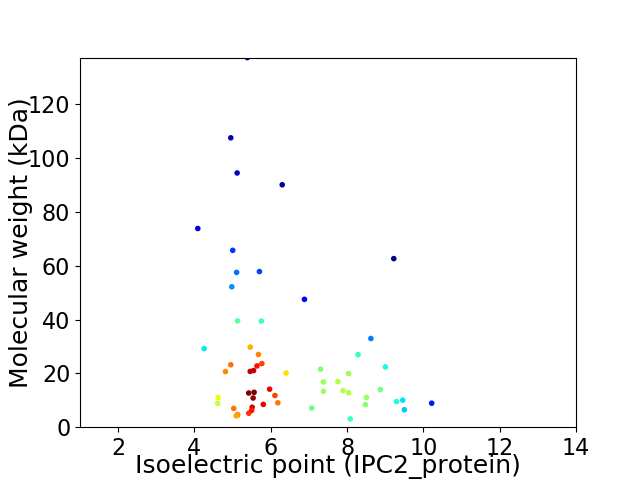
Virtual 2D-PAGE plot for 58 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
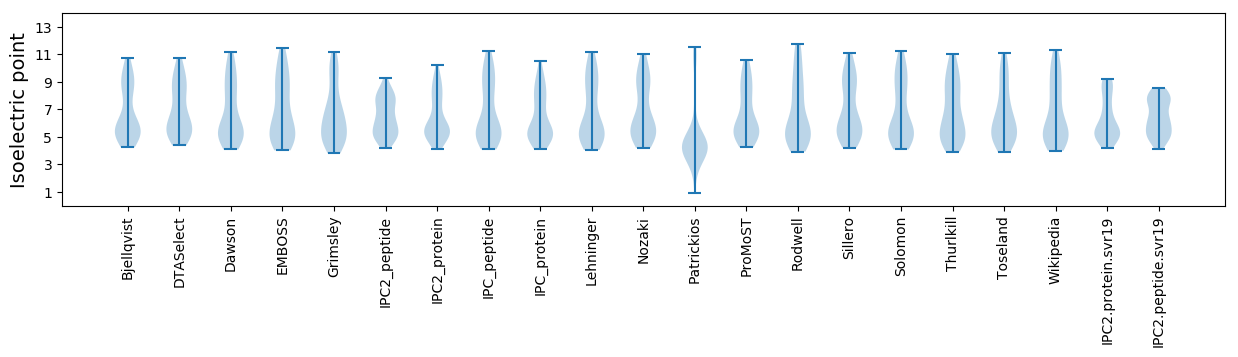
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q58N24|Q58N24_BPPRP T7-like internal core protein OS=Prochlorococcus phage P-SSP7 OX=268748 GN=16 PE=4 SV=1
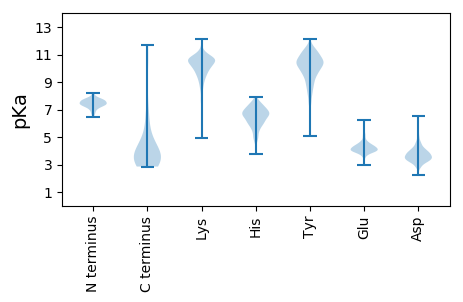
MM1 pKa = 7.35ATTEE5 pKa = 4.13HH6 pKa = 7.26FYY8 pKa = 10.66TGNGSTTSFAFTFPYY23 pKa = 9.73LANVDD28 pKa = 3.97VKK30 pKa = 11.51VEE32 pKa = 4.08LDD34 pKa = 3.42NVLKK38 pKa = 10.13TEE40 pKa = 4.11NSSGQTNNDD49 pKa = 3.22YY50 pKa = 10.45TISNTNIVFNSAPGSGVIIHH70 pKa = 7.45IYY72 pKa = 10.55RR73 pKa = 11.84NTNVDD78 pKa = 3.51TAQATYY84 pKa = 10.68AAGSSIRR91 pKa = 11.84AVDD94 pKa = 4.0LNNNQTQVLYY104 pKa = 9.38STQEE108 pKa = 3.73AQSQIIRR115 pKa = 11.84TTDD118 pKa = 2.92IKK120 pKa = 11.17DD121 pKa = 3.33GAVNSTKK128 pKa = 10.03IEE130 pKa = 4.07NNTIVNADD138 pKa = 3.63INSSAAIDD146 pKa = 3.98GSKK149 pKa = 9.75IQASSGSNSGTMSAANFTKK168 pKa = 10.63LGGIEE173 pKa = 4.05TGATADD179 pKa = 3.29QTAAEE184 pKa = 4.19IRR186 pKa = 11.84TLVEE190 pKa = 3.87SASDD194 pKa = 3.71SNVFTDD200 pKa = 4.89ADD202 pKa = 3.74HH203 pKa = 6.75TKK205 pKa = 11.05LNGIEE210 pKa = 4.22SGATADD216 pKa = 3.93QSVSEE221 pKa = 4.64IKK223 pKa = 9.84TLIAGSPLSNTHH235 pKa = 6.02IAADD239 pKa = 3.6ADD241 pKa = 3.7IAHH244 pKa = 6.66SKK246 pKa = 9.97LADD249 pKa = 3.52VTSSQVLIGNASNVPTATSLSGDD272 pKa = 3.49VTLNNAGVVTIANDD286 pKa = 3.36AVEE289 pKa = 4.65IGMIGCEE296 pKa = 3.77QTTITDD302 pKa = 3.88SDD304 pKa = 3.8SHH306 pKa = 7.77LPTSGAVVDD315 pKa = 4.31YY316 pKa = 10.88VSSQLAPIGGLEE328 pKa = 4.14VIANEE333 pKa = 4.01VSFPNTQPAAGVVISITDD351 pKa = 3.2AGGVVFNGSGSSTTGRR367 pKa = 11.84TLGGATVTINNAPTALNSEE386 pKa = 4.48TLVAGAGLMVSSTGSSNIYY405 pKa = 9.88NYY407 pKa = 10.44HH408 pKa = 6.08KK409 pKa = 10.87LFVKK413 pKa = 10.59EE414 pKa = 3.92SDD416 pKa = 3.79VVQLSDD422 pKa = 5.99DD423 pKa = 3.66INDD426 pKa = 3.2FNSRR430 pKa = 11.84YY431 pKa = 9.69RR432 pKa = 11.84IAASAPTSNNDD443 pKa = 3.14EE444 pKa = 4.01GDD446 pKa = 3.89LYY448 pKa = 11.4FDD450 pKa = 3.67TAANKK455 pKa = 9.1MFVYY459 pKa = 10.4DD460 pKa = 4.23GSAWGQVTSTGEE472 pKa = 3.91FKK474 pKa = 10.85ILGIKK479 pKa = 10.56DD480 pKa = 2.97NGQAHH485 pKa = 6.54NGSGPTFNGSNDD497 pKa = 3.32RR498 pKa = 11.84FDD500 pKa = 4.68LFEE503 pKa = 4.38NASAADD509 pKa = 3.54ISQASQLLVVLNGVIQKK526 pKa = 10.23PNDD529 pKa = 3.4GSFSGTEE536 pKa = 3.35EE537 pKa = 4.51GYY539 pKa = 11.33YY540 pKa = 10.53LDD542 pKa = 4.04GTDD545 pKa = 5.5GIRR548 pKa = 11.84FCDD551 pKa = 4.27PPASGSTLFITKK563 pKa = 9.73QGSATQINTPADD575 pKa = 3.56NTITQAKK582 pKa = 8.36IAVDD586 pKa = 3.38AVDD589 pKa = 3.7EE590 pKa = 4.25QRR592 pKa = 11.84LQVSNAPTNGYY603 pKa = 9.02YY604 pKa = 10.56LQAQSGNTGGLTWAAVPNPDD624 pKa = 3.94LTQLSASNLTSGTIPDD640 pKa = 3.53ARR642 pKa = 11.84FPSTLPAVSGANLTGLPPSCTGGGSDD668 pKa = 4.77EE669 pKa = 4.49IFWEE673 pKa = 4.17NGQTVTTNYY682 pKa = 9.98TITNNKK688 pKa = 8.86NAMSAGPITINNGIAVTIGTGEE710 pKa = 3.76NWTIVV715 pKa = 3.21
MM1 pKa = 7.35ATTEE5 pKa = 4.13HH6 pKa = 7.26FYY8 pKa = 10.66TGNGSTTSFAFTFPYY23 pKa = 9.73LANVDD28 pKa = 3.97VKK30 pKa = 11.51VEE32 pKa = 4.08LDD34 pKa = 3.42NVLKK38 pKa = 10.13TEE40 pKa = 4.11NSSGQTNNDD49 pKa = 3.22YY50 pKa = 10.45TISNTNIVFNSAPGSGVIIHH70 pKa = 7.45IYY72 pKa = 10.55RR73 pKa = 11.84NTNVDD78 pKa = 3.51TAQATYY84 pKa = 10.68AAGSSIRR91 pKa = 11.84AVDD94 pKa = 4.0LNNNQTQVLYY104 pKa = 9.38STQEE108 pKa = 3.73AQSQIIRR115 pKa = 11.84TTDD118 pKa = 2.92IKK120 pKa = 11.17DD121 pKa = 3.33GAVNSTKK128 pKa = 10.03IEE130 pKa = 4.07NNTIVNADD138 pKa = 3.63INSSAAIDD146 pKa = 3.98GSKK149 pKa = 9.75IQASSGSNSGTMSAANFTKK168 pKa = 10.63LGGIEE173 pKa = 4.05TGATADD179 pKa = 3.29QTAAEE184 pKa = 4.19IRR186 pKa = 11.84TLVEE190 pKa = 3.87SASDD194 pKa = 3.71SNVFTDD200 pKa = 4.89ADD202 pKa = 3.74HH203 pKa = 6.75TKK205 pKa = 11.05LNGIEE210 pKa = 4.22SGATADD216 pKa = 3.93QSVSEE221 pKa = 4.64IKK223 pKa = 9.84TLIAGSPLSNTHH235 pKa = 6.02IAADD239 pKa = 3.6ADD241 pKa = 3.7IAHH244 pKa = 6.66SKK246 pKa = 9.97LADD249 pKa = 3.52VTSSQVLIGNASNVPTATSLSGDD272 pKa = 3.49VTLNNAGVVTIANDD286 pKa = 3.36AVEE289 pKa = 4.65IGMIGCEE296 pKa = 3.77QTTITDD302 pKa = 3.88SDD304 pKa = 3.8SHH306 pKa = 7.77LPTSGAVVDD315 pKa = 4.31YY316 pKa = 10.88VSSQLAPIGGLEE328 pKa = 4.14VIANEE333 pKa = 4.01VSFPNTQPAAGVVISITDD351 pKa = 3.2AGGVVFNGSGSSTTGRR367 pKa = 11.84TLGGATVTINNAPTALNSEE386 pKa = 4.48TLVAGAGLMVSSTGSSNIYY405 pKa = 9.88NYY407 pKa = 10.44HH408 pKa = 6.08KK409 pKa = 10.87LFVKK413 pKa = 10.59EE414 pKa = 3.92SDD416 pKa = 3.79VVQLSDD422 pKa = 5.99DD423 pKa = 3.66INDD426 pKa = 3.2FNSRR430 pKa = 11.84YY431 pKa = 9.69RR432 pKa = 11.84IAASAPTSNNDD443 pKa = 3.14EE444 pKa = 4.01GDD446 pKa = 3.89LYY448 pKa = 11.4FDD450 pKa = 3.67TAANKK455 pKa = 9.1MFVYY459 pKa = 10.4DD460 pKa = 4.23GSAWGQVTSTGEE472 pKa = 3.91FKK474 pKa = 10.85ILGIKK479 pKa = 10.56DD480 pKa = 2.97NGQAHH485 pKa = 6.54NGSGPTFNGSNDD497 pKa = 3.32RR498 pKa = 11.84FDD500 pKa = 4.68LFEE503 pKa = 4.38NASAADD509 pKa = 3.54ISQASQLLVVLNGVIQKK526 pKa = 10.23PNDD529 pKa = 3.4GSFSGTEE536 pKa = 3.35EE537 pKa = 4.51GYY539 pKa = 11.33YY540 pKa = 10.53LDD542 pKa = 4.04GTDD545 pKa = 5.5GIRR548 pKa = 11.84FCDD551 pKa = 4.27PPASGSTLFITKK563 pKa = 9.73QGSATQINTPADD575 pKa = 3.56NTITQAKK582 pKa = 8.36IAVDD586 pKa = 3.38AVDD589 pKa = 3.7EE590 pKa = 4.25QRR592 pKa = 11.84LQVSNAPTNGYY603 pKa = 9.02YY604 pKa = 10.56LQAQSGNTGGLTWAAVPNPDD624 pKa = 3.94LTQLSASNLTSGTIPDD640 pKa = 3.53ARR642 pKa = 11.84FPSTLPAVSGANLTGLPPSCTGGGSDD668 pKa = 4.77EE669 pKa = 4.49IFWEE673 pKa = 4.17NGQTVTTNYY682 pKa = 9.98TITNNKK688 pKa = 8.86NAMSAGPITINNGIAVTIGTGEE710 pKa = 3.76NWTIVV715 pKa = 3.21
Molecular weight: 73.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q58N12|Q58N12_BPPRP Gp49 OS=Prochlorococcus phage P-SSP7 OX=268748 GN=PCPG_00046 PE=4 SV=1
MM1 pKa = 7.75AGGAKK6 pKa = 9.35YY7 pKa = 10.99ANGNYY12 pKa = 9.8KK13 pKa = 9.95SQQKK17 pKa = 10.55AYY19 pKa = 10.0NKK21 pKa = 8.3TKK23 pKa = 10.35KK24 pKa = 10.57GLALRR29 pKa = 11.84VNANAINRR37 pKa = 11.84AKK39 pKa = 9.59GTYY42 pKa = 10.39GNGDD46 pKa = 3.76GKK48 pKa = 10.61DD49 pKa = 3.45VAHH52 pKa = 7.32KK53 pKa = 10.43KK54 pKa = 10.32GKK56 pKa = 9.93QNSKK60 pKa = 9.46KK61 pKa = 10.8AKK63 pKa = 10.0DD64 pKa = 3.46ATLQSPSKK72 pKa = 10.23NRR74 pKa = 11.84KK75 pKa = 8.58SRR77 pKa = 11.84LKK79 pKa = 10.06IRR81 pKa = 11.84KK82 pKa = 8.67
MM1 pKa = 7.75AGGAKK6 pKa = 9.35YY7 pKa = 10.99ANGNYY12 pKa = 9.8KK13 pKa = 9.95SQQKK17 pKa = 10.55AYY19 pKa = 10.0NKK21 pKa = 8.3TKK23 pKa = 10.35KK24 pKa = 10.57GLALRR29 pKa = 11.84VNANAINRR37 pKa = 11.84AKK39 pKa = 9.59GTYY42 pKa = 10.39GNGDD46 pKa = 3.76GKK48 pKa = 10.61DD49 pKa = 3.45VAHH52 pKa = 7.32KK53 pKa = 10.43KK54 pKa = 10.32GKK56 pKa = 9.93QNSKK60 pKa = 9.46KK61 pKa = 10.8AKK63 pKa = 10.0DD64 pKa = 3.46ATLQSPSKK72 pKa = 10.23NRR74 pKa = 11.84KK75 pKa = 8.58SRR77 pKa = 11.84LKK79 pKa = 10.06IRR81 pKa = 11.84KK82 pKa = 8.67
Molecular weight: 8.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14200 |
26 |
1245 |
244.8 |
27.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.113 ± 0.383 | 1.113 ± 0.172 |
6.148 ± 0.242 | 6.113 ± 0.396 |
3.711 ± 0.175 | 6.803 ± 0.36 |
1.775 ± 0.2 | 6.042 ± 0.214 |
6.859 ± 0.569 | 8.197 ± 0.316 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.239 ± 0.161 | 5.387 ± 0.368 |
4.148 ± 0.234 | 4.204 ± 0.21 |
4.451 ± 0.264 | 6.803 ± 0.401 |
7.211 ± 0.484 | 5.901 ± 0.307 |
1.303 ± 0.109 | 3.479 ± 0.171 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
