
Sanxia water strider virus 14
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.75
Get precalculated fractions of proteins
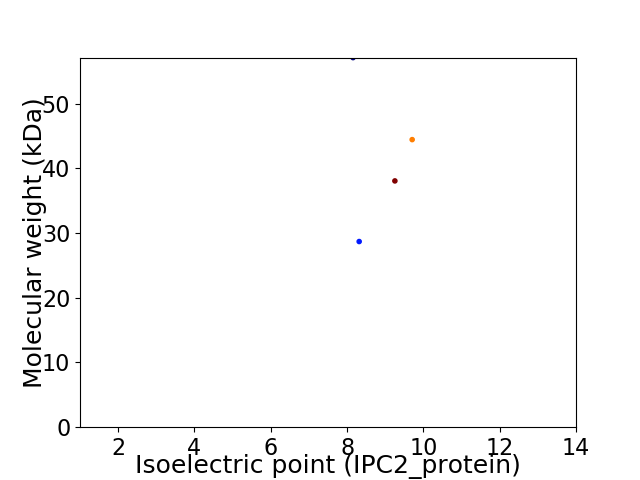
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
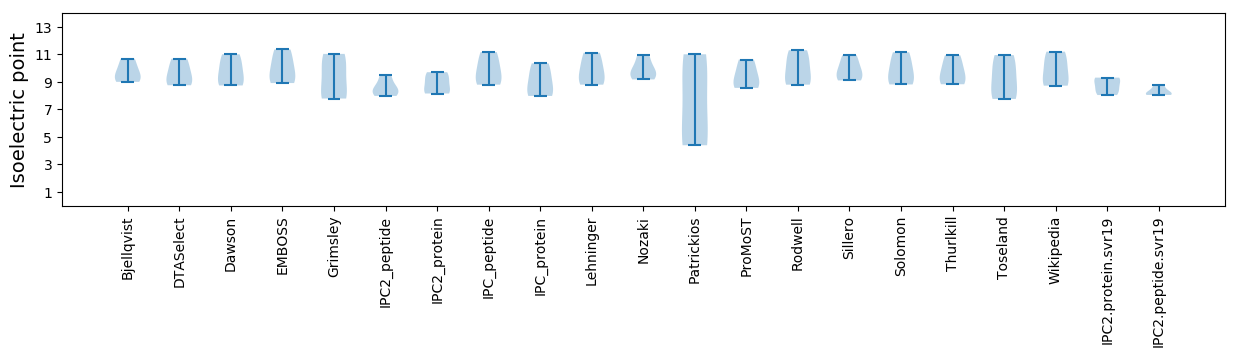
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGJ6|A0A1L3KGJ6_9VIRU RNA-directed RNA polymerase OS=Sanxia water strider virus 14 OX=1923398 PE=4 SV=1
MM1 pKa = 7.69PKK3 pKa = 10.33SKK5 pKa = 10.05GHH7 pKa = 6.27SLLPNCSHH15 pKa = 7.29RR16 pKa = 11.84AKK18 pKa = 10.25TSDD21 pKa = 3.75CRR23 pKa = 11.84CSRR26 pKa = 11.84ATYY29 pKa = 10.94KK30 pKa = 10.96LFDD33 pKa = 3.78YY34 pKa = 10.92SLEE37 pKa = 4.07HH38 pKa = 7.11LNEE41 pKa = 4.29SVVWTHH47 pKa = 6.97RR48 pKa = 11.84GCVCNEE54 pKa = 3.62LVALRR59 pKa = 11.84EE60 pKa = 3.94RR61 pKa = 11.84HH62 pKa = 5.55QLDD65 pKa = 2.92TGARR69 pKa = 11.84YY70 pKa = 8.9SAPINAVSVNVDD82 pKa = 3.61EE83 pKa = 6.05LVMANAGRR91 pKa = 11.84FPEE94 pKa = 4.39LKK96 pKa = 9.92RR97 pKa = 11.84NAFSVVISHH106 pKa = 6.03YY107 pKa = 10.84SGGKK111 pKa = 7.33RR112 pKa = 11.84AEE114 pKa = 3.98YY115 pKa = 10.19EE116 pKa = 3.88RR117 pKa = 11.84ARR119 pKa = 11.84EE120 pKa = 3.98SLLIDD125 pKa = 3.75PLVEE129 pKa = 4.1RR130 pKa = 11.84DD131 pKa = 3.0SRR133 pKa = 11.84LRR135 pKa = 11.84MFLKK139 pKa = 10.36DD140 pKa = 4.09DD141 pKa = 5.01KK142 pKa = 11.17YY143 pKa = 10.68HH144 pKa = 5.67TWDD147 pKa = 4.47FKK149 pKa = 11.37APRR152 pKa = 11.84CIQFRR157 pKa = 11.84SKK159 pKa = 10.76RR160 pKa = 11.84YY161 pKa = 9.24GLTLSSYY168 pKa = 8.91LQPIEE173 pKa = 4.04EE174 pKa = 4.25FTYY177 pKa = 10.4EE178 pKa = 4.88LRR180 pKa = 11.84DD181 pKa = 3.66TTDD184 pKa = 2.46SHH186 pKa = 7.71VFAKK190 pKa = 10.37SRR192 pKa = 11.84NLDD195 pKa = 3.25DD196 pKa = 4.07RR197 pKa = 11.84AADD200 pKa = 5.32LRR202 pKa = 11.84LKK204 pKa = 9.51WDD206 pKa = 3.6SFIDD210 pKa = 4.51PIAYY214 pKa = 10.24CLDD217 pKa = 3.32HH218 pKa = 7.19SKK220 pKa = 10.62FDD222 pKa = 3.53CHH224 pKa = 7.73VNVDD228 pKa = 4.23LLKK231 pKa = 10.48QEE233 pKa = 3.73HH234 pKa = 6.52RR235 pKa = 11.84YY236 pKa = 9.35YY237 pKa = 10.54RR238 pKa = 11.84QFYY241 pKa = 9.65PRR243 pKa = 11.84DD244 pKa = 3.28AGLRR248 pKa = 11.84KK249 pKa = 9.67LLSQQINNRR258 pKa = 11.84GSTKK262 pKa = 10.77NGTTYY267 pKa = 8.78KK268 pKa = 9.6TCGTRR273 pKa = 11.84MSGDD277 pKa = 3.73PNTGLGNSILNYY289 pKa = 10.48GMLRR293 pKa = 11.84EE294 pKa = 4.11AFKK297 pKa = 10.69KK298 pKa = 10.29VRR300 pKa = 11.84AAYY303 pKa = 10.3YY304 pKa = 9.78IDD306 pKa = 5.19GDD308 pKa = 3.96DD309 pKa = 3.39SVVIIEE315 pKa = 5.04SADD318 pKa = 3.46QVNVNVGCFEE328 pKa = 4.23RR329 pKa = 11.84FGMKK333 pKa = 9.6TKK335 pKa = 10.82VEE337 pKa = 3.93VATIFEE343 pKa = 4.39HH344 pKa = 7.35TEE346 pKa = 3.87FCQCRR351 pKa = 11.84PVYY354 pKa = 10.59DD355 pKa = 3.57GLKK358 pKa = 7.98WHH360 pKa = 6.97LVRR363 pKa = 11.84NPYY366 pKa = 8.39RR367 pKa = 11.84TLARR371 pKa = 11.84LPWIVKK377 pKa = 9.88KK378 pKa = 10.64NHH380 pKa = 6.39LSCKK384 pKa = 9.52ARR386 pKa = 11.84YY387 pKa = 8.13IKK389 pKa = 10.71SVGLCEE395 pKa = 4.24LALNMGIPVLQQIASLMVEE414 pKa = 4.05RR415 pKa = 11.84GEE417 pKa = 4.19GKK419 pKa = 10.16YY420 pKa = 10.38IKK422 pKa = 9.52TDD424 pKa = 2.89RR425 pKa = 11.84HH426 pKa = 5.47FMAKK430 pKa = 9.95RR431 pKa = 11.84ATIKK435 pKa = 9.6PWHH438 pKa = 6.21ARR440 pKa = 11.84KK441 pKa = 10.07VPIRR445 pKa = 11.84EE446 pKa = 3.88VTRR449 pKa = 11.84EE450 pKa = 3.94SFEE453 pKa = 4.16IAWGISRR460 pKa = 11.84EE461 pKa = 4.06EE462 pKa = 3.78QLEE465 pKa = 4.31LEE467 pKa = 4.53NVTLSLPHH475 pKa = 6.94GDD477 pKa = 4.09AEE479 pKa = 4.8SLFYY483 pKa = 10.35THH485 pKa = 7.62LAPGVTHH492 pKa = 6.6CEE494 pKa = 4.24CC495 pKa = 5.1
MM1 pKa = 7.69PKK3 pKa = 10.33SKK5 pKa = 10.05GHH7 pKa = 6.27SLLPNCSHH15 pKa = 7.29RR16 pKa = 11.84AKK18 pKa = 10.25TSDD21 pKa = 3.75CRR23 pKa = 11.84CSRR26 pKa = 11.84ATYY29 pKa = 10.94KK30 pKa = 10.96LFDD33 pKa = 3.78YY34 pKa = 10.92SLEE37 pKa = 4.07HH38 pKa = 7.11LNEE41 pKa = 4.29SVVWTHH47 pKa = 6.97RR48 pKa = 11.84GCVCNEE54 pKa = 3.62LVALRR59 pKa = 11.84EE60 pKa = 3.94RR61 pKa = 11.84HH62 pKa = 5.55QLDD65 pKa = 2.92TGARR69 pKa = 11.84YY70 pKa = 8.9SAPINAVSVNVDD82 pKa = 3.61EE83 pKa = 6.05LVMANAGRR91 pKa = 11.84FPEE94 pKa = 4.39LKK96 pKa = 9.92RR97 pKa = 11.84NAFSVVISHH106 pKa = 6.03YY107 pKa = 10.84SGGKK111 pKa = 7.33RR112 pKa = 11.84AEE114 pKa = 3.98YY115 pKa = 10.19EE116 pKa = 3.88RR117 pKa = 11.84ARR119 pKa = 11.84EE120 pKa = 3.98SLLIDD125 pKa = 3.75PLVEE129 pKa = 4.1RR130 pKa = 11.84DD131 pKa = 3.0SRR133 pKa = 11.84LRR135 pKa = 11.84MFLKK139 pKa = 10.36DD140 pKa = 4.09DD141 pKa = 5.01KK142 pKa = 11.17YY143 pKa = 10.68HH144 pKa = 5.67TWDD147 pKa = 4.47FKK149 pKa = 11.37APRR152 pKa = 11.84CIQFRR157 pKa = 11.84SKK159 pKa = 10.76RR160 pKa = 11.84YY161 pKa = 9.24GLTLSSYY168 pKa = 8.91LQPIEE173 pKa = 4.04EE174 pKa = 4.25FTYY177 pKa = 10.4EE178 pKa = 4.88LRR180 pKa = 11.84DD181 pKa = 3.66TTDD184 pKa = 2.46SHH186 pKa = 7.71VFAKK190 pKa = 10.37SRR192 pKa = 11.84NLDD195 pKa = 3.25DD196 pKa = 4.07RR197 pKa = 11.84AADD200 pKa = 5.32LRR202 pKa = 11.84LKK204 pKa = 9.51WDD206 pKa = 3.6SFIDD210 pKa = 4.51PIAYY214 pKa = 10.24CLDD217 pKa = 3.32HH218 pKa = 7.19SKK220 pKa = 10.62FDD222 pKa = 3.53CHH224 pKa = 7.73VNVDD228 pKa = 4.23LLKK231 pKa = 10.48QEE233 pKa = 3.73HH234 pKa = 6.52RR235 pKa = 11.84YY236 pKa = 9.35YY237 pKa = 10.54RR238 pKa = 11.84QFYY241 pKa = 9.65PRR243 pKa = 11.84DD244 pKa = 3.28AGLRR248 pKa = 11.84KK249 pKa = 9.67LLSQQINNRR258 pKa = 11.84GSTKK262 pKa = 10.77NGTTYY267 pKa = 8.78KK268 pKa = 9.6TCGTRR273 pKa = 11.84MSGDD277 pKa = 3.73PNTGLGNSILNYY289 pKa = 10.48GMLRR293 pKa = 11.84EE294 pKa = 4.11AFKK297 pKa = 10.69KK298 pKa = 10.29VRR300 pKa = 11.84AAYY303 pKa = 10.3YY304 pKa = 9.78IDD306 pKa = 5.19GDD308 pKa = 3.96DD309 pKa = 3.39SVVIIEE315 pKa = 5.04SADD318 pKa = 3.46QVNVNVGCFEE328 pKa = 4.23RR329 pKa = 11.84FGMKK333 pKa = 9.6TKK335 pKa = 10.82VEE337 pKa = 3.93VATIFEE343 pKa = 4.39HH344 pKa = 7.35TEE346 pKa = 3.87FCQCRR351 pKa = 11.84PVYY354 pKa = 10.59DD355 pKa = 3.57GLKK358 pKa = 7.98WHH360 pKa = 6.97LVRR363 pKa = 11.84NPYY366 pKa = 8.39RR367 pKa = 11.84TLARR371 pKa = 11.84LPWIVKK377 pKa = 9.88KK378 pKa = 10.64NHH380 pKa = 6.39LSCKK384 pKa = 9.52ARR386 pKa = 11.84YY387 pKa = 8.13IKK389 pKa = 10.71SVGLCEE395 pKa = 4.24LALNMGIPVLQQIASLMVEE414 pKa = 4.05RR415 pKa = 11.84GEE417 pKa = 4.19GKK419 pKa = 10.16YY420 pKa = 10.38IKK422 pKa = 9.52TDD424 pKa = 2.89RR425 pKa = 11.84HH426 pKa = 5.47FMAKK430 pKa = 9.95RR431 pKa = 11.84ATIKK435 pKa = 9.6PWHH438 pKa = 6.21ARR440 pKa = 11.84KK441 pKa = 10.07VPIRR445 pKa = 11.84EE446 pKa = 3.88VTRR449 pKa = 11.84EE450 pKa = 3.94SFEE453 pKa = 4.16IAWGISRR460 pKa = 11.84EE461 pKa = 4.06EE462 pKa = 3.78QLEE465 pKa = 4.31LEE467 pKa = 4.53NVTLSLPHH475 pKa = 6.94GDD477 pKa = 4.09AEE479 pKa = 4.8SLFYY483 pKa = 10.35THH485 pKa = 7.62LAPGVTHH492 pKa = 6.6CEE494 pKa = 4.24CC495 pKa = 5.1
Molecular weight: 57.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG71|A0A1L3KG71_9VIRU Uncharacterized protein OS=Sanxia water strider virus 14 OX=1923398 PE=4 SV=1
MM1 pKa = 6.44QTSTSYY7 pKa = 11.23KK8 pKa = 9.62PDD10 pKa = 3.12LKK12 pKa = 11.1KK13 pKa = 10.91FGEE16 pKa = 4.15QRR18 pKa = 11.84ARR20 pKa = 11.84TSWQGPKK27 pKa = 10.04SSQNGFRR34 pKa = 11.84RR35 pKa = 11.84TPTGQGSPAANKK47 pKa = 7.32QQCSSCGEE55 pKa = 3.78LGKK58 pKa = 10.4RR59 pKa = 11.84LRR61 pKa = 11.84KK62 pKa = 9.77LEE64 pKa = 3.79EE65 pKa = 4.43RR66 pKa = 11.84IRR68 pKa = 11.84TLSNKK73 pKa = 9.26HH74 pKa = 5.03VRR76 pKa = 11.84LPSATGSWQTVCRR89 pKa = 11.84EE90 pKa = 3.59KK91 pKa = 10.83VAPKK95 pKa = 10.05SRR97 pKa = 11.84QRR99 pKa = 11.84PTTSHH104 pKa = 6.06QDD106 pKa = 3.11RR107 pKa = 11.84TGVKK111 pKa = 9.49PLNNQEE117 pKa = 3.84RR118 pKa = 11.84DD119 pKa = 3.56TILRR123 pKa = 11.84KK124 pKa = 9.45HH125 pKa = 6.69DD126 pKa = 4.26LLNPSSTNQARR137 pKa = 11.84RR138 pKa = 11.84VGSVPNVSSAGTSGEE153 pKa = 4.31SMTPLLKK160 pKa = 9.69TASLSRR166 pKa = 11.84ILPSRR171 pKa = 11.84RR172 pKa = 11.84TSPRR176 pKa = 11.84RR177 pKa = 11.84SSRR180 pKa = 11.84RR181 pKa = 11.84SDD183 pKa = 2.74ATTSAKK189 pKa = 10.24KK190 pKa = 10.65SSGPDD195 pKa = 3.29AACATAPRR203 pKa = 11.84TAKK206 pKa = 10.37AAAATVQSGSVIEE219 pKa = 4.79ALATTPKK226 pKa = 10.31DD227 pKa = 3.49LKK229 pKa = 10.59RR230 pKa = 11.84RR231 pKa = 11.84PGYY234 pKa = 10.46KK235 pKa = 9.02SDD237 pKa = 3.96RR238 pKa = 11.84DD239 pKa = 3.62SPTIKK244 pKa = 10.18RR245 pKa = 11.84VTWEE249 pKa = 3.66ARR251 pKa = 11.84RR252 pKa = 11.84APDD255 pKa = 3.79DD256 pKa = 4.4AADD259 pKa = 3.76KK260 pKa = 10.49PVYY263 pKa = 9.08ATNTSKK269 pKa = 10.84RR270 pKa = 11.84GPARR274 pKa = 11.84KK275 pKa = 9.5VPLGEE280 pKa = 4.01RR281 pKa = 11.84RR282 pKa = 11.84RR283 pKa = 11.84QRR285 pKa = 11.84KK286 pKa = 9.17AFVDD290 pKa = 3.59VDD292 pKa = 4.46DD293 pKa = 4.08NLYY296 pKa = 9.89WYY298 pKa = 10.52LVLEE302 pKa = 4.68FALVPRR308 pKa = 11.84TSDD311 pKa = 2.79ILRR314 pKa = 11.84QMKK317 pKa = 10.33AKK319 pKa = 10.42AKK321 pKa = 10.28QFLNLHH327 pKa = 6.46DD328 pKa = 5.64LNDD331 pKa = 3.62ISLKK335 pKa = 10.57DD336 pKa = 3.53QYY338 pKa = 11.46TMVIGTISRR347 pKa = 11.84AMLVPEE353 pKa = 4.19QEE355 pKa = 3.79MKK357 pKa = 10.64LRR359 pKa = 11.84QEE361 pKa = 4.59LKK363 pKa = 9.63SDD365 pKa = 3.45AAIEE369 pKa = 4.09EE370 pKa = 4.42MGKK373 pKa = 9.14HH374 pKa = 5.36AKK376 pKa = 9.16LVKK379 pKa = 10.02RR380 pKa = 11.84GMIGRR385 pKa = 11.84AFGGLGKK392 pKa = 9.76PFSLPEE398 pKa = 4.47AIKK401 pKa = 10.82
MM1 pKa = 6.44QTSTSYY7 pKa = 11.23KK8 pKa = 9.62PDD10 pKa = 3.12LKK12 pKa = 11.1KK13 pKa = 10.91FGEE16 pKa = 4.15QRR18 pKa = 11.84ARR20 pKa = 11.84TSWQGPKK27 pKa = 10.04SSQNGFRR34 pKa = 11.84RR35 pKa = 11.84TPTGQGSPAANKK47 pKa = 7.32QQCSSCGEE55 pKa = 3.78LGKK58 pKa = 10.4RR59 pKa = 11.84LRR61 pKa = 11.84KK62 pKa = 9.77LEE64 pKa = 3.79EE65 pKa = 4.43RR66 pKa = 11.84IRR68 pKa = 11.84TLSNKK73 pKa = 9.26HH74 pKa = 5.03VRR76 pKa = 11.84LPSATGSWQTVCRR89 pKa = 11.84EE90 pKa = 3.59KK91 pKa = 10.83VAPKK95 pKa = 10.05SRR97 pKa = 11.84QRR99 pKa = 11.84PTTSHH104 pKa = 6.06QDD106 pKa = 3.11RR107 pKa = 11.84TGVKK111 pKa = 9.49PLNNQEE117 pKa = 3.84RR118 pKa = 11.84DD119 pKa = 3.56TILRR123 pKa = 11.84KK124 pKa = 9.45HH125 pKa = 6.69DD126 pKa = 4.26LLNPSSTNQARR137 pKa = 11.84RR138 pKa = 11.84VGSVPNVSSAGTSGEE153 pKa = 4.31SMTPLLKK160 pKa = 9.69TASLSRR166 pKa = 11.84ILPSRR171 pKa = 11.84RR172 pKa = 11.84TSPRR176 pKa = 11.84RR177 pKa = 11.84SSRR180 pKa = 11.84RR181 pKa = 11.84SDD183 pKa = 2.74ATTSAKK189 pKa = 10.24KK190 pKa = 10.65SSGPDD195 pKa = 3.29AACATAPRR203 pKa = 11.84TAKK206 pKa = 10.37AAAATVQSGSVIEE219 pKa = 4.79ALATTPKK226 pKa = 10.31DD227 pKa = 3.49LKK229 pKa = 10.59RR230 pKa = 11.84RR231 pKa = 11.84PGYY234 pKa = 10.46KK235 pKa = 9.02SDD237 pKa = 3.96RR238 pKa = 11.84DD239 pKa = 3.62SPTIKK244 pKa = 10.18RR245 pKa = 11.84VTWEE249 pKa = 3.66ARR251 pKa = 11.84RR252 pKa = 11.84APDD255 pKa = 3.79DD256 pKa = 4.4AADD259 pKa = 3.76KK260 pKa = 10.49PVYY263 pKa = 9.08ATNTSKK269 pKa = 10.84RR270 pKa = 11.84GPARR274 pKa = 11.84KK275 pKa = 9.5VPLGEE280 pKa = 4.01RR281 pKa = 11.84RR282 pKa = 11.84RR283 pKa = 11.84QRR285 pKa = 11.84KK286 pKa = 9.17AFVDD290 pKa = 3.59VDD292 pKa = 4.46DD293 pKa = 4.08NLYY296 pKa = 9.89WYY298 pKa = 10.52LVLEE302 pKa = 4.68FALVPRR308 pKa = 11.84TSDD311 pKa = 2.79ILRR314 pKa = 11.84QMKK317 pKa = 10.33AKK319 pKa = 10.42AKK321 pKa = 10.28QFLNLHH327 pKa = 6.46DD328 pKa = 5.64LNDD331 pKa = 3.62ISLKK335 pKa = 10.57DD336 pKa = 3.53QYY338 pKa = 11.46TMVIGTISRR347 pKa = 11.84AMLVPEE353 pKa = 4.19QEE355 pKa = 3.79MKK357 pKa = 10.64LRR359 pKa = 11.84QEE361 pKa = 4.59LKK363 pKa = 9.63SDD365 pKa = 3.45AAIEE369 pKa = 4.09EE370 pKa = 4.42MGKK373 pKa = 9.14HH374 pKa = 5.36AKK376 pKa = 9.16LVKK379 pKa = 10.02RR380 pKa = 11.84GMIGRR385 pKa = 11.84AFGGLGKK392 pKa = 9.76PFSLPEE398 pKa = 4.47AIKK401 pKa = 10.82
Molecular weight: 44.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1503 |
254 |
495 |
375.8 |
42.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.051 ± 0.617 | 2.129 ± 0.739 |
4.657 ± 0.482 | 5.522 ± 0.78 |
3.061 ± 0.428 | 6.587 ± 0.809 |
2.262 ± 0.785 | 4.125 ± 0.621 |
6.188 ± 0.855 | 7.585 ± 0.824 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.063 ± 0.351 | 3.992 ± 0.278 |
5.522 ± 0.762 | 3.859 ± 0.589 |
9.115 ± 0.699 | 7.984 ± 0.6 |
7.319 ± 1.332 | 5.921 ± 0.55 |
1.264 ± 0.197 | 2.794 ± 0.645 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
