
Enterobacteria phage IME10
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; Lederbergvirus; unclassified Lederbergvirus
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
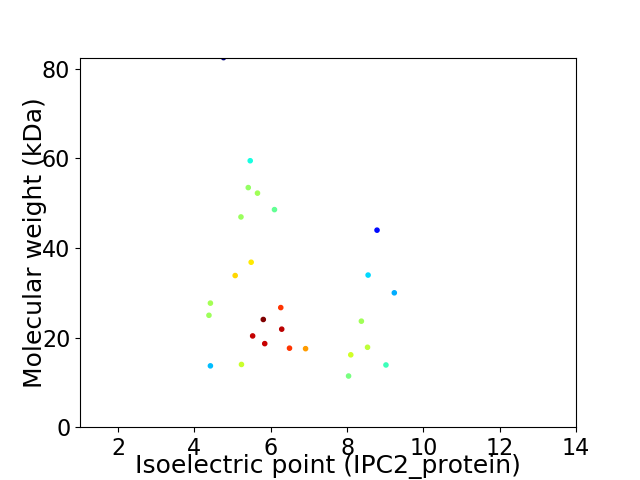
Virtual 2D-PAGE plot for 27 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G5DA86|G5DA86_9CAUD ORF252 OS=Enterobacteria phage IME10 OX=1090133 PE=4 SV=1
MM1 pKa = 7.7ASMKK5 pKa = 9.68IDD7 pKa = 3.55YY8 pKa = 9.66QALRR12 pKa = 11.84EE13 pKa = 4.09KK14 pKa = 10.94AEE16 pKa = 4.07KK17 pKa = 8.76ATCGVWSLEE26 pKa = 4.05YY27 pKa = 11.02GEE29 pKa = 5.26SRR31 pKa = 11.84FDD33 pKa = 3.96GDD35 pKa = 3.49DD36 pKa = 3.43ALIHH40 pKa = 6.59RR41 pKa = 11.84EE42 pKa = 3.79VAGYY46 pKa = 10.15IPICRR51 pKa = 11.84IEE53 pKa = 4.3GAHH56 pKa = 6.67PEE58 pKa = 4.21SGFDD62 pKa = 3.18EE63 pKa = 4.95DD64 pKa = 5.53FQIEE68 pKa = 4.14QQANAEE74 pKa = 4.43FIAAANTATVLALLGEE90 pKa = 4.45LEE92 pKa = 4.32TAKK95 pKa = 10.59KK96 pKa = 10.32RR97 pKa = 11.84IAEE100 pKa = 4.18LEE102 pKa = 4.22AEE104 pKa = 4.66PVSQTCKK111 pKa = 10.62LPVNTPCQDD120 pKa = 3.39APSHH124 pKa = 6.03IWLQTAGVWPEE135 pKa = 4.02DD136 pKa = 3.88GEE138 pKa = 4.48LSEE141 pKa = 5.49LTWCSHH147 pKa = 5.17NQHH150 pKa = 6.77HH151 pKa = 8.11DD152 pKa = 3.11DD153 pKa = 3.69TLYY156 pKa = 11.06VRR158 pKa = 11.84ADD160 pKa = 3.67LVNGNSPGTPDD171 pKa = 2.86SWISCSEE178 pKa = 3.88RR179 pKa = 11.84MPDD182 pKa = 3.77SNCLYY187 pKa = 10.02LCWGTYY193 pKa = 9.72FEE195 pKa = 5.52GDD197 pKa = 3.54EE198 pKa = 4.23PDD200 pKa = 4.18YY201 pKa = 11.1IPAYY205 pKa = 10.33FFVHH209 pKa = 5.91KK210 pKa = 9.68TNGWTEE216 pKa = 3.95WQPVEE221 pKa = 5.09DD222 pKa = 4.16DD223 pKa = 3.8CNPRR227 pKa = 11.84EE228 pKa = 4.17VIITHH233 pKa = 6.41WMPLPAAPKK242 pKa = 10.22QEE244 pKa = 4.11VKK246 pKa = 10.84
MM1 pKa = 7.7ASMKK5 pKa = 9.68IDD7 pKa = 3.55YY8 pKa = 9.66QALRR12 pKa = 11.84EE13 pKa = 4.09KK14 pKa = 10.94AEE16 pKa = 4.07KK17 pKa = 8.76ATCGVWSLEE26 pKa = 4.05YY27 pKa = 11.02GEE29 pKa = 5.26SRR31 pKa = 11.84FDD33 pKa = 3.96GDD35 pKa = 3.49DD36 pKa = 3.43ALIHH40 pKa = 6.59RR41 pKa = 11.84EE42 pKa = 3.79VAGYY46 pKa = 10.15IPICRR51 pKa = 11.84IEE53 pKa = 4.3GAHH56 pKa = 6.67PEE58 pKa = 4.21SGFDD62 pKa = 3.18EE63 pKa = 4.95DD64 pKa = 5.53FQIEE68 pKa = 4.14QQANAEE74 pKa = 4.43FIAAANTATVLALLGEE90 pKa = 4.45LEE92 pKa = 4.32TAKK95 pKa = 10.59KK96 pKa = 10.32RR97 pKa = 11.84IAEE100 pKa = 4.18LEE102 pKa = 4.22AEE104 pKa = 4.66PVSQTCKK111 pKa = 10.62LPVNTPCQDD120 pKa = 3.39APSHH124 pKa = 6.03IWLQTAGVWPEE135 pKa = 4.02DD136 pKa = 3.88GEE138 pKa = 4.48LSEE141 pKa = 5.49LTWCSHH147 pKa = 5.17NQHH150 pKa = 6.77HH151 pKa = 8.11DD152 pKa = 3.11DD153 pKa = 3.69TLYY156 pKa = 11.06VRR158 pKa = 11.84ADD160 pKa = 3.67LVNGNSPGTPDD171 pKa = 2.86SWISCSEE178 pKa = 3.88RR179 pKa = 11.84MPDD182 pKa = 3.77SNCLYY187 pKa = 10.02LCWGTYY193 pKa = 9.72FEE195 pKa = 5.52GDD197 pKa = 3.54EE198 pKa = 4.23PDD200 pKa = 4.18YY201 pKa = 11.1IPAYY205 pKa = 10.33FFVHH209 pKa = 5.91KK210 pKa = 9.68TNGWTEE216 pKa = 3.95WQPVEE221 pKa = 5.09DD222 pKa = 4.16DD223 pKa = 3.8CNPRR227 pKa = 11.84EE228 pKa = 4.17VIITHH233 pKa = 6.41WMPLPAAPKK242 pKa = 10.22QEE244 pKa = 4.11VKK246 pKa = 10.84
Molecular weight: 27.69 kDa
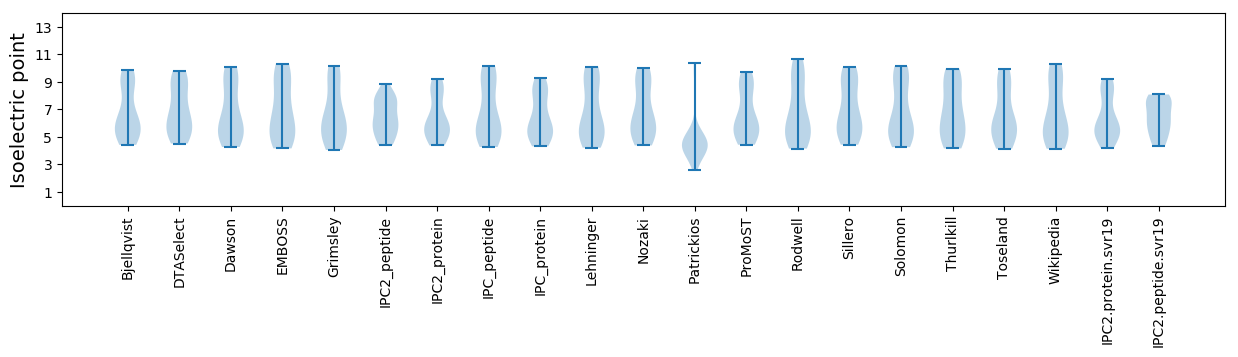
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8EYI7|G8EYI7_9CAUD Terminase small subunit OS=Enterobacteria phage IME10 OX=1090133 GN=3 PE=4 SV=1
MM1 pKa = 7.75EE2 pKa = 5.76PRR4 pKa = 11.84TNQQLTRR11 pKa = 11.84RR12 pKa = 11.84DD13 pKa = 3.15RR14 pKa = 11.84EE15 pKa = 3.84RR16 pKa = 11.84NQRR19 pKa = 11.84RR20 pKa = 11.84GHH22 pKa = 4.2VHH24 pKa = 5.74LWSYY28 pKa = 11.44ASSEE32 pKa = 4.33TTQICQPDD40 pKa = 4.01VYY42 pKa = 10.13RR43 pKa = 11.84YY44 pKa = 9.56RR45 pKa = 11.84SSVLRR50 pKa = 11.84KK51 pKa = 8.29PHH53 pKa = 6.63IDD55 pKa = 3.06SGTGRR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84MRR64 pKa = 11.84LEE66 pKa = 3.91SVAKK70 pKa = 9.15FHH72 pKa = 6.87SPKK75 pKa = 10.44SPMMSDD81 pKa = 3.41SPRR84 pKa = 11.84ATASDD89 pKa = 4.01SLSGTDD95 pKa = 3.38VMAAMGMAQSQAGFGMAAFCGKK117 pKa = 9.93HH118 pKa = 5.73EE119 pKa = 4.72LSQNDD124 pKa = 3.28KK125 pKa = 10.89QKK127 pKa = 11.16AINYY131 pKa = 8.5LMQFAHH137 pKa = 6.59KK138 pKa = 10.66VSGKK142 pKa = 9.17YY143 pKa = 9.64RR144 pKa = 11.84GVAKK148 pKa = 10.96LEE150 pKa = 4.3GNTKK154 pKa = 10.64AKK156 pKa = 10.08VLQVLATFAYY166 pKa = 10.49ADD168 pKa = 3.87YY169 pKa = 9.9CRR171 pKa = 11.84SAATPGARR179 pKa = 11.84CRR181 pKa = 11.84DD182 pKa = 3.55CHH184 pKa = 5.14GTGRR188 pKa = 11.84AVDD191 pKa = 3.51IAKK194 pKa = 8.9TEE196 pKa = 3.51QWGRR200 pKa = 11.84VVEE203 pKa = 4.53KK204 pKa = 10.74EE205 pKa = 3.88CGRR208 pKa = 11.84CKK210 pKa = 10.51GVGYY214 pKa = 10.85SRR216 pKa = 11.84MPASAAYY223 pKa = 9.71RR224 pKa = 11.84AVTMLIPNLTQPTWSRR240 pKa = 11.84TVKK243 pKa = 10.04PLYY246 pKa = 10.17DD247 pKa = 3.46ALVVQCHH254 pKa = 5.63KK255 pKa = 10.78EE256 pKa = 3.65EE257 pKa = 4.83SIADD261 pKa = 3.94NILNAVTRR269 pKa = 4.15
MM1 pKa = 7.75EE2 pKa = 5.76PRR4 pKa = 11.84TNQQLTRR11 pKa = 11.84RR12 pKa = 11.84DD13 pKa = 3.15RR14 pKa = 11.84EE15 pKa = 3.84RR16 pKa = 11.84NQRR19 pKa = 11.84RR20 pKa = 11.84GHH22 pKa = 4.2VHH24 pKa = 5.74LWSYY28 pKa = 11.44ASSEE32 pKa = 4.33TTQICQPDD40 pKa = 4.01VYY42 pKa = 10.13RR43 pKa = 11.84YY44 pKa = 9.56RR45 pKa = 11.84SSVLRR50 pKa = 11.84KK51 pKa = 8.29PHH53 pKa = 6.63IDD55 pKa = 3.06SGTGRR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84MRR64 pKa = 11.84LEE66 pKa = 3.91SVAKK70 pKa = 9.15FHH72 pKa = 6.87SPKK75 pKa = 10.44SPMMSDD81 pKa = 3.41SPRR84 pKa = 11.84ATASDD89 pKa = 4.01SLSGTDD95 pKa = 3.38VMAAMGMAQSQAGFGMAAFCGKK117 pKa = 9.93HH118 pKa = 5.73EE119 pKa = 4.72LSQNDD124 pKa = 3.28KK125 pKa = 10.89QKK127 pKa = 11.16AINYY131 pKa = 8.5LMQFAHH137 pKa = 6.59KK138 pKa = 10.66VSGKK142 pKa = 9.17YY143 pKa = 9.64RR144 pKa = 11.84GVAKK148 pKa = 10.96LEE150 pKa = 4.3GNTKK154 pKa = 10.64AKK156 pKa = 10.08VLQVLATFAYY166 pKa = 10.49ADD168 pKa = 3.87YY169 pKa = 9.9CRR171 pKa = 11.84SAATPGARR179 pKa = 11.84CRR181 pKa = 11.84DD182 pKa = 3.55CHH184 pKa = 5.14GTGRR188 pKa = 11.84AVDD191 pKa = 3.51IAKK194 pKa = 8.9TEE196 pKa = 3.51QWGRR200 pKa = 11.84VVEE203 pKa = 4.53KK204 pKa = 10.74EE205 pKa = 3.88CGRR208 pKa = 11.84CKK210 pKa = 10.51GVGYY214 pKa = 10.85SRR216 pKa = 11.84MPASAAYY223 pKa = 9.71RR224 pKa = 11.84AVTMLIPNLTQPTWSRR240 pKa = 11.84TVKK243 pKa = 10.04PLYY246 pKa = 10.17DD247 pKa = 3.46ALVVQCHH254 pKa = 5.63KK255 pKa = 10.78EE256 pKa = 3.65EE257 pKa = 4.83SIADD261 pKa = 3.94NILNAVTRR269 pKa = 4.15
Molecular weight: 29.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7427 |
107 |
721 |
275.1 |
30.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.465 ± 0.655 | 1.118 ± 0.177 |
6.247 ± 0.265 | 7.136 ± 0.747 |
2.976 ± 0.231 | 6.907 ± 0.424 |
1.616 ± 0.169 | 5.695 ± 0.271 |
6.14 ± 0.341 | 7.54 ± 0.317 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.895 ± 0.188 | 4.753 ± 0.344 |
3.932 ± 0.297 | 5.426 ± 0.467 |
6.099 ± 0.381 | 6.019 ± 0.417 |
5.17 ± 0.331 | 5.924 ± 0.456 |
1.548 ± 0.213 | 3.393 ± 0.279 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
