
Asterias forbesi associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; unclassified Circovirus
Average proteome isoelectric point is 8.56
Get precalculated fractions of proteins
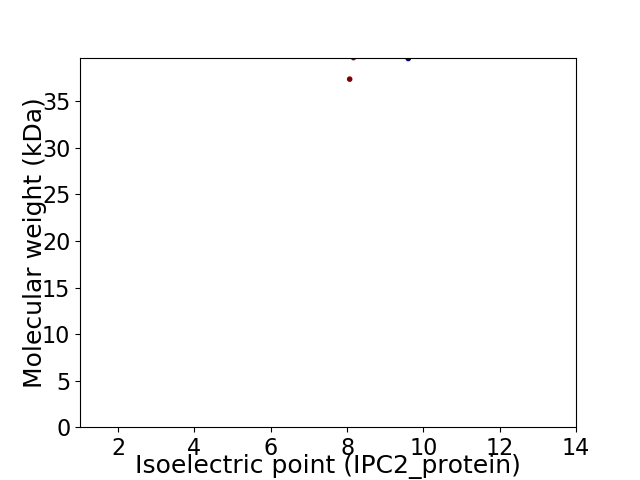
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H4FV52|A0A0H4FV52_9CIRC Putative capsid protein OS=Asterias forbesi associated circular virus OX=1676988 PE=4 SV=1
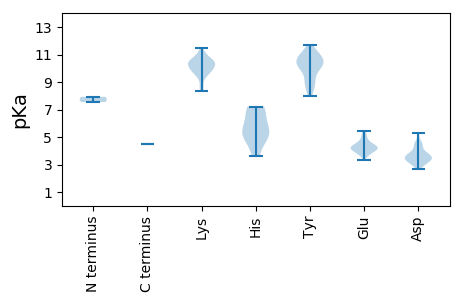
MM1 pKa = 7.93KK2 pKa = 10.16SRR4 pKa = 11.84NWIFTINNEE13 pKa = 4.84LIWKK17 pKa = 8.84PEE19 pKa = 3.32NHH21 pKa = 6.89LIKK24 pKa = 10.59FVVYY28 pKa = 10.11QLEE31 pKa = 4.14EE32 pKa = 4.34GEE34 pKa = 4.44KK35 pKa = 10.37EE36 pKa = 4.23KK37 pKa = 11.18VPHH40 pKa = 5.29IQGYY44 pKa = 10.18LEE46 pKa = 4.46LKK48 pKa = 9.95QPRR51 pKa = 11.84ALSWMKK57 pKa = 10.66KK58 pKa = 9.87LNSKK62 pKa = 9.41AHH64 pKa = 5.01WEE66 pKa = 3.78IRR68 pKa = 11.84KK69 pKa = 9.7GNRR72 pKa = 11.84VQALQYY78 pKa = 7.97VTKK81 pKa = 10.59EE82 pKa = 4.09EE83 pKa = 4.14TRR85 pKa = 11.84MDD87 pKa = 3.87APWILNCGQWSQYY100 pKa = 10.43DD101 pKa = 3.28ASTWTRR107 pKa = 11.84VEE109 pKa = 3.89DD110 pKa = 3.56WLNTVIPGVWKK121 pKa = 10.65DD122 pKa = 3.6GSKK125 pKa = 8.59STKK128 pKa = 10.02SIRR131 pKa = 11.84LSEE134 pKa = 4.26IKK136 pKa = 10.6SKK138 pKa = 11.1LDD140 pKa = 3.81DD141 pKa = 4.29GCSSEE146 pKa = 5.43VIADD150 pKa = 3.62NYY152 pKa = 9.98FEE154 pKa = 4.8DD155 pKa = 3.62WVRR158 pKa = 11.84HH159 pKa = 3.64YY160 pKa = 10.89RR161 pKa = 11.84AFEE164 pKa = 3.98KK165 pKa = 11.09YY166 pKa = 10.52LLIKK170 pKa = 10.31SKK172 pKa = 10.44PRR174 pKa = 11.84DD175 pKa = 3.4YY176 pKa = 10.38PVRR179 pKa = 11.84VVVVIGPTGTGKK191 pKa = 10.38SRR193 pKa = 11.84FCMEE197 pKa = 4.71QYY199 pKa = 10.85PDD201 pKa = 4.6AYY203 pKa = 9.31WKK205 pKa = 10.09QRR207 pKa = 11.84SQWWDD212 pKa = 3.42GYY214 pKa = 9.88VRR216 pKa = 11.84HH217 pKa = 5.82KK218 pKa = 9.83TIILDD223 pKa = 4.02EE224 pKa = 4.48YY225 pKa = 10.88YY226 pKa = 11.01GWLPFDD232 pKa = 4.48LLLRR236 pKa = 11.84LCDD239 pKa = 4.37RR240 pKa = 11.84YY241 pKa = 10.68PLMLEE246 pKa = 4.36TKK248 pKa = 10.34GGQCNCEE255 pKa = 3.87ANTIVITSNNRR266 pKa = 11.84PDD268 pKa = 3.19SWYY271 pKa = 10.39KK272 pKa = 10.1NCYY275 pKa = 8.81FPAFEE280 pKa = 4.34RR281 pKa = 11.84RR282 pKa = 11.84VDD284 pKa = 3.18TFIIMKK290 pKa = 9.59EE291 pKa = 3.98LGSSFIFKK299 pKa = 9.88KK300 pKa = 9.94WSDD303 pKa = 3.79AEE305 pKa = 4.17KK306 pKa = 9.33HH307 pKa = 5.35TDD309 pKa = 3.61PLILNN314 pKa = 4.52
MM1 pKa = 7.93KK2 pKa = 10.16SRR4 pKa = 11.84NWIFTINNEE13 pKa = 4.84LIWKK17 pKa = 8.84PEE19 pKa = 3.32NHH21 pKa = 6.89LIKK24 pKa = 10.59FVVYY28 pKa = 10.11QLEE31 pKa = 4.14EE32 pKa = 4.34GEE34 pKa = 4.44KK35 pKa = 10.37EE36 pKa = 4.23KK37 pKa = 11.18VPHH40 pKa = 5.29IQGYY44 pKa = 10.18LEE46 pKa = 4.46LKK48 pKa = 9.95QPRR51 pKa = 11.84ALSWMKK57 pKa = 10.66KK58 pKa = 9.87LNSKK62 pKa = 9.41AHH64 pKa = 5.01WEE66 pKa = 3.78IRR68 pKa = 11.84KK69 pKa = 9.7GNRR72 pKa = 11.84VQALQYY78 pKa = 7.97VTKK81 pKa = 10.59EE82 pKa = 4.09EE83 pKa = 4.14TRR85 pKa = 11.84MDD87 pKa = 3.87APWILNCGQWSQYY100 pKa = 10.43DD101 pKa = 3.28ASTWTRR107 pKa = 11.84VEE109 pKa = 3.89DD110 pKa = 3.56WLNTVIPGVWKK121 pKa = 10.65DD122 pKa = 3.6GSKK125 pKa = 8.59STKK128 pKa = 10.02SIRR131 pKa = 11.84LSEE134 pKa = 4.26IKK136 pKa = 10.6SKK138 pKa = 11.1LDD140 pKa = 3.81DD141 pKa = 4.29GCSSEE146 pKa = 5.43VIADD150 pKa = 3.62NYY152 pKa = 9.98FEE154 pKa = 4.8DD155 pKa = 3.62WVRR158 pKa = 11.84HH159 pKa = 3.64YY160 pKa = 10.89RR161 pKa = 11.84AFEE164 pKa = 3.98KK165 pKa = 11.09YY166 pKa = 10.52LLIKK170 pKa = 10.31SKK172 pKa = 10.44PRR174 pKa = 11.84DD175 pKa = 3.4YY176 pKa = 10.38PVRR179 pKa = 11.84VVVVIGPTGTGKK191 pKa = 10.38SRR193 pKa = 11.84FCMEE197 pKa = 4.71QYY199 pKa = 10.85PDD201 pKa = 4.6AYY203 pKa = 9.31WKK205 pKa = 10.09QRR207 pKa = 11.84SQWWDD212 pKa = 3.42GYY214 pKa = 9.88VRR216 pKa = 11.84HH217 pKa = 5.82KK218 pKa = 9.83TIILDD223 pKa = 4.02EE224 pKa = 4.48YY225 pKa = 10.88YY226 pKa = 11.01GWLPFDD232 pKa = 4.48LLLRR236 pKa = 11.84LCDD239 pKa = 4.37RR240 pKa = 11.84YY241 pKa = 10.68PLMLEE246 pKa = 4.36TKK248 pKa = 10.34GGQCNCEE255 pKa = 3.87ANTIVITSNNRR266 pKa = 11.84PDD268 pKa = 3.19SWYY271 pKa = 10.39KK272 pKa = 10.1NCYY275 pKa = 8.81FPAFEE280 pKa = 4.34RR281 pKa = 11.84RR282 pKa = 11.84VDD284 pKa = 3.18TFIIMKK290 pKa = 9.59EE291 pKa = 3.98LGSSFIFKK299 pKa = 9.88KK300 pKa = 9.94WSDD303 pKa = 3.79AEE305 pKa = 4.17KK306 pKa = 9.33HH307 pKa = 5.35TDD309 pKa = 3.61PLILNN314 pKa = 4.52
Molecular weight: 37.39 kDa
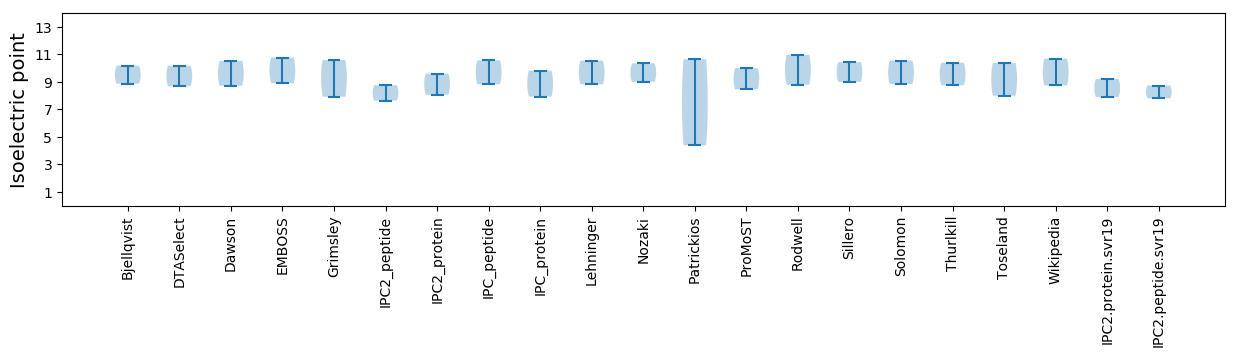
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H4FV52|A0A0H4FV52_9CIRC Putative capsid protein OS=Asterias forbesi associated circular virus OX=1676988 PE=4 SV=1
MM1 pKa = 7.54PRR3 pKa = 11.84NTRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84TGRR11 pKa = 11.84SRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84TLRR19 pKa = 11.84RR20 pKa = 11.84GYY22 pKa = 10.67RR23 pKa = 11.84KK24 pKa = 8.34FTGGRR29 pKa = 11.84GITKK33 pKa = 9.77EE34 pKa = 3.6HH35 pKa = 7.21DD36 pKa = 3.22RR37 pKa = 11.84TNIYY41 pKa = 9.25YY42 pKa = 10.13KK43 pKa = 10.7KK44 pKa = 10.37RR45 pKa = 11.84MPRR48 pKa = 11.84FKK50 pKa = 10.23KK51 pKa = 9.92RR52 pKa = 11.84RR53 pKa = 11.84WRR55 pKa = 11.84RR56 pKa = 11.84FSKK59 pKa = 10.3KK60 pKa = 8.4VQNVSEE66 pKa = 4.66KK67 pKa = 10.99DD68 pKa = 3.21LGTQTVVFNDD78 pKa = 3.62QIEE81 pKa = 4.35TGDD84 pKa = 3.79FFNSDD89 pKa = 3.41GLITGSATLCLYY101 pKa = 11.08SMNGSEE107 pKa = 5.18VPYY110 pKa = 10.94NDD112 pKa = 4.5LAVMMNNLNFAAPTVDD128 pKa = 5.26LGTTKK133 pKa = 10.47YY134 pKa = 8.25RR135 pKa = 11.84TSKK138 pKa = 10.95VIFKK142 pKa = 10.67SAVFDD147 pKa = 3.61LTLTNTSYY155 pKa = 11.67DD156 pKa = 3.61RR157 pKa = 11.84IGGVNNIEE165 pKa = 3.83AAYY168 pKa = 9.88SLEE171 pKa = 4.1VDD173 pKa = 2.92IHH175 pKa = 5.07TVVIKK180 pKa = 10.71KK181 pKa = 9.39GARR184 pKa = 11.84DD185 pKa = 3.35LDD187 pKa = 4.64FISGTLAEE195 pKa = 5.18KK196 pKa = 10.56GLKK199 pKa = 9.56TIDD202 pKa = 3.39KK203 pKa = 8.95MIEE206 pKa = 3.86VAEE209 pKa = 4.25KK210 pKa = 9.6EE211 pKa = 4.11VRR213 pKa = 11.84PIGGDD218 pKa = 3.16DD219 pKa = 3.42SKK221 pKa = 11.49IFNTTKK227 pKa = 10.72AKK229 pKa = 10.03FNRR232 pKa = 11.84GGTLWDD238 pKa = 3.69VPHH241 pKa = 6.87ALSLFGMKK249 pKa = 9.74ILSKK253 pKa = 10.42KK254 pKa = 10.36KK255 pKa = 9.55YY256 pKa = 8.96FIPGGATITYY266 pKa = 8.86QMRR269 pKa = 11.84DD270 pKa = 2.87AKK272 pKa = 10.36RR273 pKa = 11.84RR274 pKa = 11.84VMVPQFNMLGVNKK287 pKa = 10.08PGWTKK292 pKa = 10.4FLYY295 pKa = 8.68ITYY298 pKa = 10.17KK299 pKa = 10.39LVPGVGVIGQDD310 pKa = 3.29EE311 pKa = 4.27NVPTQIRR318 pKa = 11.84TRR320 pKa = 11.84LSVGVTRR327 pKa = 11.84KK328 pKa = 9.94YY329 pKa = 10.24SYY331 pKa = 10.61KK332 pKa = 10.05IEE334 pKa = 4.29GQNDD338 pKa = 2.67VRR340 pKa = 11.84DD341 pKa = 4.1NYY343 pKa = 11.34NPDD346 pKa = 3.03LL347 pKa = 4.52
MM1 pKa = 7.54PRR3 pKa = 11.84NTRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84TGRR11 pKa = 11.84SRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84TLRR19 pKa = 11.84RR20 pKa = 11.84GYY22 pKa = 10.67RR23 pKa = 11.84KK24 pKa = 8.34FTGGRR29 pKa = 11.84GITKK33 pKa = 9.77EE34 pKa = 3.6HH35 pKa = 7.21DD36 pKa = 3.22RR37 pKa = 11.84TNIYY41 pKa = 9.25YY42 pKa = 10.13KK43 pKa = 10.7KK44 pKa = 10.37RR45 pKa = 11.84MPRR48 pKa = 11.84FKK50 pKa = 10.23KK51 pKa = 9.92RR52 pKa = 11.84RR53 pKa = 11.84WRR55 pKa = 11.84RR56 pKa = 11.84FSKK59 pKa = 10.3KK60 pKa = 8.4VQNVSEE66 pKa = 4.66KK67 pKa = 10.99DD68 pKa = 3.21LGTQTVVFNDD78 pKa = 3.62QIEE81 pKa = 4.35TGDD84 pKa = 3.79FFNSDD89 pKa = 3.41GLITGSATLCLYY101 pKa = 11.08SMNGSEE107 pKa = 5.18VPYY110 pKa = 10.94NDD112 pKa = 4.5LAVMMNNLNFAAPTVDD128 pKa = 5.26LGTTKK133 pKa = 10.47YY134 pKa = 8.25RR135 pKa = 11.84TSKK138 pKa = 10.95VIFKK142 pKa = 10.67SAVFDD147 pKa = 3.61LTLTNTSYY155 pKa = 11.67DD156 pKa = 3.61RR157 pKa = 11.84IGGVNNIEE165 pKa = 3.83AAYY168 pKa = 9.88SLEE171 pKa = 4.1VDD173 pKa = 2.92IHH175 pKa = 5.07TVVIKK180 pKa = 10.71KK181 pKa = 9.39GARR184 pKa = 11.84DD185 pKa = 3.35LDD187 pKa = 4.64FISGTLAEE195 pKa = 5.18KK196 pKa = 10.56GLKK199 pKa = 9.56TIDD202 pKa = 3.39KK203 pKa = 8.95MIEE206 pKa = 3.86VAEE209 pKa = 4.25KK210 pKa = 9.6EE211 pKa = 4.11VRR213 pKa = 11.84PIGGDD218 pKa = 3.16DD219 pKa = 3.42SKK221 pKa = 11.49IFNTTKK227 pKa = 10.72AKK229 pKa = 10.03FNRR232 pKa = 11.84GGTLWDD238 pKa = 3.69VPHH241 pKa = 6.87ALSLFGMKK249 pKa = 9.74ILSKK253 pKa = 10.42KK254 pKa = 10.36KK255 pKa = 9.55YY256 pKa = 8.96FIPGGATITYY266 pKa = 8.86QMRR269 pKa = 11.84DD270 pKa = 2.87AKK272 pKa = 10.36RR273 pKa = 11.84RR274 pKa = 11.84VMVPQFNMLGVNKK287 pKa = 10.08PGWTKK292 pKa = 10.4FLYY295 pKa = 8.68ITYY298 pKa = 10.17KK299 pKa = 10.39LVPGVGVIGQDD310 pKa = 3.29EE311 pKa = 4.27NVPTQIRR318 pKa = 11.84TRR320 pKa = 11.84LSVGVTRR327 pKa = 11.84KK328 pKa = 9.94YY329 pKa = 10.24SYY331 pKa = 10.61KK332 pKa = 10.05IEE334 pKa = 4.29GQNDD338 pKa = 2.67VRR340 pKa = 11.84DD341 pKa = 4.1NYY343 pKa = 11.34NPDD346 pKa = 3.03LL347 pKa = 4.52
Molecular weight: 39.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
661 |
314 |
347 |
330.5 |
38.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.782 ± 0.191 | 1.21 ± 0.699 |
5.9 ± 0.115 | 5.144 ± 1.278 |
4.085 ± 0.399 | 6.808 ± 1.394 |
1.362 ± 0.377 | 6.354 ± 0.229 |
8.926 ± 0.006 | 7.262 ± 0.48 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.421 ± 0.35 | 5.446 ± 0.459 |
3.933 ± 0.36 | 2.874 ± 0.432 |
7.716 ± 1.142 | 5.598 ± 0.53 |
6.959 ± 1.498 | 6.657 ± 0.634 |
2.874 ± 1.524 | 4.69 ± 0.278 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
