
Rodent Torque teno virus 2
Taxonomy: Viruses; Anelloviridae; unclassified Anelloviridae
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
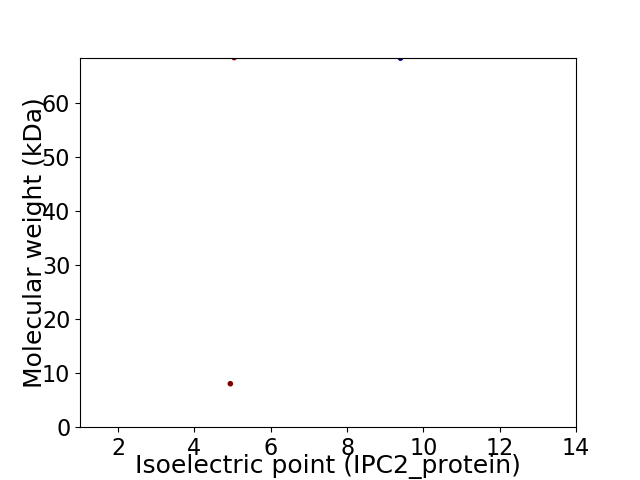
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|X2G4Y3|X2G4Y3_9VIRU Capsid protein OS=Rodent Torque teno virus 2 OX=1514665 PE=3 SV=1
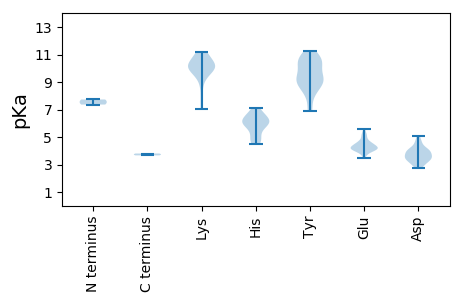
MM1 pKa = 7.36NPSILAFKK9 pKa = 9.81QQEE12 pKa = 4.04ALFRR16 pKa = 11.84QAVRR20 pKa = 11.84AQHH23 pKa = 7.11RR24 pKa = 11.84LFCDD28 pKa = 3.31CGDD31 pKa = 3.97PVQHH35 pKa = 6.6LLGWRR40 pKa = 11.84TSTGGPFVTLAGTTTGEE57 pKa = 4.16DD58 pKa = 3.22TGEE61 pKa = 4.33GGSMEE66 pKa = 5.0LDD68 pKa = 3.6GEE70 pKa = 4.52HH71 pKa = 5.89TAEE74 pKa = 4.23GG75 pKa = 3.67
MM1 pKa = 7.36NPSILAFKK9 pKa = 9.81QQEE12 pKa = 4.04ALFRR16 pKa = 11.84QAVRR20 pKa = 11.84AQHH23 pKa = 7.11RR24 pKa = 11.84LFCDD28 pKa = 3.31CGDD31 pKa = 3.97PVQHH35 pKa = 6.6LLGWRR40 pKa = 11.84TSTGGPFVTLAGTTTGEE57 pKa = 4.16DD58 pKa = 3.22TGEE61 pKa = 4.33GGSMEE66 pKa = 5.0LDD68 pKa = 3.6GEE70 pKa = 4.52HH71 pKa = 5.89TAEE74 pKa = 4.23GG75 pKa = 3.67
Molecular weight: 8.03 kDa
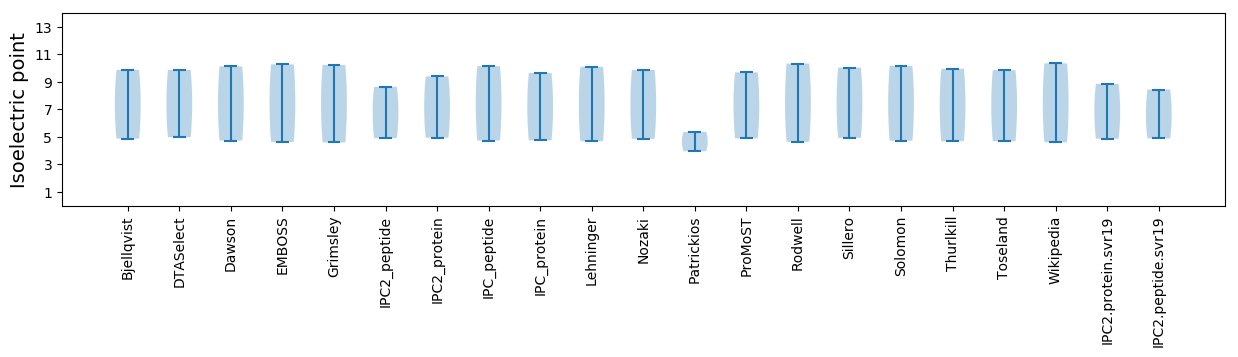
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|X2G4Y3|X2G4Y3_9VIRU Capsid protein OS=Rodent Torque teno virus 2 OX=1514665 PE=3 SV=1
MM1 pKa = 7.73AYY3 pKa = 10.08FYY5 pKa = 10.77RR6 pKa = 11.84RR7 pKa = 11.84TLRR10 pKa = 11.84YY11 pKa = 8.78PRR13 pKa = 11.84RR14 pKa = 11.84NYY16 pKa = 9.21YY17 pKa = 9.02RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84YY21 pKa = 8.94RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84QYY27 pKa = 8.08GTRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84TYY34 pKa = 8.4RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84IRR39 pKa = 11.84RR40 pKa = 11.84PGKK43 pKa = 7.01WRR45 pKa = 11.84RR46 pKa = 11.84YY47 pKa = 6.96YY48 pKa = 9.03RR49 pKa = 11.84QYY51 pKa = 11.22SRR53 pKa = 11.84RR54 pKa = 11.84HH55 pKa = 4.72AAKK58 pKa = 9.72IVQWNPAVRR67 pKa = 11.84TKK69 pKa = 10.88CKK71 pKa = 10.27IIGLTPLMMALGADD85 pKa = 3.55SGIQDD90 pKa = 5.04FILPTTSAVVLWKK103 pKa = 10.91GGGVDD108 pKa = 4.52SGTLSLLDD116 pKa = 4.63LYY118 pKa = 10.49WEE120 pKa = 4.12EE121 pKa = 4.25RR122 pKa = 11.84FWRR125 pKa = 11.84ARR127 pKa = 11.84WSKK130 pKa = 11.21SNQGYY135 pKa = 8.65NLCRR139 pKa = 11.84YY140 pKa = 9.5FGVKK144 pKa = 8.62LTLYY148 pKa = 10.17RR149 pKa = 11.84LQHH152 pKa = 5.28HH153 pKa = 6.67TYY155 pKa = 9.07IFWWTVEE162 pKa = 4.72DD163 pKa = 4.09FTDD166 pKa = 3.9DD167 pKa = 3.82QEE169 pKa = 4.59PLTVCHH175 pKa = 6.81PSQLLLTRR183 pKa = 11.84NHH185 pKa = 6.12VLVKK189 pKa = 10.25HH190 pKa = 5.81YY191 pKa = 10.82GSNRR195 pKa = 11.84QVDD198 pKa = 3.47KK199 pKa = 11.0AVRR202 pKa = 11.84IKK204 pKa = 10.51IPPPTYY210 pKa = 10.49LDD212 pKa = 4.12SSWHH216 pKa = 4.78TFKK219 pKa = 11.14DD220 pKa = 3.36IAKK223 pKa = 9.98KK224 pKa = 10.13PLLKK228 pKa = 9.99WRR230 pKa = 11.84ASLIDD235 pKa = 4.08TEE237 pKa = 4.72NPWTGYY243 pKa = 9.02PDD245 pKa = 3.29NRR247 pKa = 11.84VGLRR251 pKa = 11.84IYY253 pKa = 10.36LWSWDD258 pKa = 3.63TATKK262 pKa = 9.67TGKK265 pKa = 9.21QAEE268 pKa = 4.49VMYY271 pKa = 9.13FPPLDD276 pKa = 4.04EE277 pKa = 5.11GNNTQICIHH286 pKa = 6.21KK287 pKa = 10.37ADD289 pKa = 4.13WNNEE293 pKa = 3.78GTGPKK298 pKa = 9.45QDD300 pKa = 4.73AANFWPDD307 pKa = 3.41RR308 pKa = 11.84NDD310 pKa = 2.84RR311 pKa = 11.84WEE313 pKa = 4.07TLVVPFYY320 pKa = 10.67MFTFGRR326 pKa = 11.84NAEE329 pKa = 4.06WYY331 pKa = 6.9DD332 pKa = 3.92TNRR335 pKa = 11.84HH336 pKa = 4.46RR337 pKa = 11.84HH338 pKa = 4.9LPKK341 pKa = 10.3PEE343 pKa = 4.13PEE345 pKa = 4.01GNMGYY350 pKa = 10.52FLYY353 pKa = 10.73VKK355 pKa = 10.31HH356 pKa = 6.45NSNPAWRR363 pKa = 11.84DD364 pKa = 3.0GVTGQLITTWEE375 pKa = 4.3MAFMTYY381 pKa = 8.11NTAQRR386 pKa = 11.84IAASGPWVEE395 pKa = 5.56KK396 pKa = 10.64GIQAPGINVTMRR408 pKa = 11.84YY409 pKa = 9.62SFQFQWGGRR418 pKa = 11.84PGTKK422 pKa = 10.06LPPEE426 pKa = 4.66APDD429 pKa = 3.0TGGPHH434 pKa = 6.05WPKK437 pKa = 9.97ATYY440 pKa = 9.56RR441 pKa = 11.84WPGSLRR447 pKa = 11.84ADD449 pKa = 2.74IRR451 pKa = 11.84DD452 pKa = 3.57PTEE455 pKa = 3.76INHH458 pKa = 6.19EE459 pKa = 4.22VLQGEE464 pKa = 5.14DD465 pKa = 3.79YY466 pKa = 10.87DD467 pKa = 4.22SSGIITEE474 pKa = 4.23GAFRR478 pKa = 11.84RR479 pKa = 11.84VAKK482 pKa = 9.37PHH484 pKa = 6.23YY485 pKa = 9.87ISTDD489 pKa = 3.1EE490 pKa = 5.26KK491 pKa = 9.95DD492 pKa = 3.94WIWGRR497 pKa = 11.84LRR499 pKa = 11.84HH500 pKa = 5.97PPPSKK505 pKa = 10.29RR506 pKa = 11.84KK507 pKa = 7.81RR508 pKa = 11.84QRR510 pKa = 11.84PSSEE514 pKa = 3.49EE515 pKa = 4.05RR516 pKa = 11.84EE517 pKa = 4.11TDD519 pKa = 3.43GSKK522 pKa = 10.91EE523 pKa = 4.45DD524 pKa = 3.53YY525 pKa = 9.18TQASEE530 pKa = 4.55TEE532 pKa = 4.46TEE534 pKa = 4.32TEE536 pKa = 4.13TEE538 pKa = 4.11DD539 pKa = 3.71EE540 pKa = 4.28LGHH543 pKa = 5.65RR544 pKa = 11.84RR545 pKa = 11.84KK546 pKa = 10.11RR547 pKa = 11.84NKK549 pKa = 10.08LRR551 pKa = 11.84ILLDD555 pKa = 3.4RR556 pKa = 11.84LEE558 pKa = 4.37IRR560 pKa = 11.84PHH562 pKa = 6.08ILTNRR567 pKa = 11.84SGNWEE572 pKa = 4.05SASII576 pKa = 3.8
MM1 pKa = 7.73AYY3 pKa = 10.08FYY5 pKa = 10.77RR6 pKa = 11.84RR7 pKa = 11.84TLRR10 pKa = 11.84YY11 pKa = 8.78PRR13 pKa = 11.84RR14 pKa = 11.84NYY16 pKa = 9.21YY17 pKa = 9.02RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84YY21 pKa = 8.94RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84QYY27 pKa = 8.08GTRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84TYY34 pKa = 8.4RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84IRR39 pKa = 11.84RR40 pKa = 11.84PGKK43 pKa = 7.01WRR45 pKa = 11.84RR46 pKa = 11.84YY47 pKa = 6.96YY48 pKa = 9.03RR49 pKa = 11.84QYY51 pKa = 11.22SRR53 pKa = 11.84RR54 pKa = 11.84HH55 pKa = 4.72AAKK58 pKa = 9.72IVQWNPAVRR67 pKa = 11.84TKK69 pKa = 10.88CKK71 pKa = 10.27IIGLTPLMMALGADD85 pKa = 3.55SGIQDD90 pKa = 5.04FILPTTSAVVLWKK103 pKa = 10.91GGGVDD108 pKa = 4.52SGTLSLLDD116 pKa = 4.63LYY118 pKa = 10.49WEE120 pKa = 4.12EE121 pKa = 4.25RR122 pKa = 11.84FWRR125 pKa = 11.84ARR127 pKa = 11.84WSKK130 pKa = 11.21SNQGYY135 pKa = 8.65NLCRR139 pKa = 11.84YY140 pKa = 9.5FGVKK144 pKa = 8.62LTLYY148 pKa = 10.17RR149 pKa = 11.84LQHH152 pKa = 5.28HH153 pKa = 6.67TYY155 pKa = 9.07IFWWTVEE162 pKa = 4.72DD163 pKa = 4.09FTDD166 pKa = 3.9DD167 pKa = 3.82QEE169 pKa = 4.59PLTVCHH175 pKa = 6.81PSQLLLTRR183 pKa = 11.84NHH185 pKa = 6.12VLVKK189 pKa = 10.25HH190 pKa = 5.81YY191 pKa = 10.82GSNRR195 pKa = 11.84QVDD198 pKa = 3.47KK199 pKa = 11.0AVRR202 pKa = 11.84IKK204 pKa = 10.51IPPPTYY210 pKa = 10.49LDD212 pKa = 4.12SSWHH216 pKa = 4.78TFKK219 pKa = 11.14DD220 pKa = 3.36IAKK223 pKa = 9.98KK224 pKa = 10.13PLLKK228 pKa = 9.99WRR230 pKa = 11.84ASLIDD235 pKa = 4.08TEE237 pKa = 4.72NPWTGYY243 pKa = 9.02PDD245 pKa = 3.29NRR247 pKa = 11.84VGLRR251 pKa = 11.84IYY253 pKa = 10.36LWSWDD258 pKa = 3.63TATKK262 pKa = 9.67TGKK265 pKa = 9.21QAEE268 pKa = 4.49VMYY271 pKa = 9.13FPPLDD276 pKa = 4.04EE277 pKa = 5.11GNNTQICIHH286 pKa = 6.21KK287 pKa = 10.37ADD289 pKa = 4.13WNNEE293 pKa = 3.78GTGPKK298 pKa = 9.45QDD300 pKa = 4.73AANFWPDD307 pKa = 3.41RR308 pKa = 11.84NDD310 pKa = 2.84RR311 pKa = 11.84WEE313 pKa = 4.07TLVVPFYY320 pKa = 10.67MFTFGRR326 pKa = 11.84NAEE329 pKa = 4.06WYY331 pKa = 6.9DD332 pKa = 3.92TNRR335 pKa = 11.84HH336 pKa = 4.46RR337 pKa = 11.84HH338 pKa = 4.9LPKK341 pKa = 10.3PEE343 pKa = 4.13PEE345 pKa = 4.01GNMGYY350 pKa = 10.52FLYY353 pKa = 10.73VKK355 pKa = 10.31HH356 pKa = 6.45NSNPAWRR363 pKa = 11.84DD364 pKa = 3.0GVTGQLITTWEE375 pKa = 4.3MAFMTYY381 pKa = 8.11NTAQRR386 pKa = 11.84IAASGPWVEE395 pKa = 5.56KK396 pKa = 10.64GIQAPGINVTMRR408 pKa = 11.84YY409 pKa = 9.62SFQFQWGGRR418 pKa = 11.84PGTKK422 pKa = 10.06LPPEE426 pKa = 4.66APDD429 pKa = 3.0TGGPHH434 pKa = 6.05WPKK437 pKa = 9.97ATYY440 pKa = 9.56RR441 pKa = 11.84WPGSLRR447 pKa = 11.84ADD449 pKa = 2.74IRR451 pKa = 11.84DD452 pKa = 3.57PTEE455 pKa = 3.76INHH458 pKa = 6.19EE459 pKa = 4.22VLQGEE464 pKa = 5.14DD465 pKa = 3.79YY466 pKa = 10.87DD467 pKa = 4.22SSGIITEE474 pKa = 4.23GAFRR478 pKa = 11.84RR479 pKa = 11.84VAKK482 pKa = 9.37PHH484 pKa = 6.23YY485 pKa = 9.87ISTDD489 pKa = 3.1EE490 pKa = 5.26KK491 pKa = 9.95DD492 pKa = 3.94WIWGRR497 pKa = 11.84LRR499 pKa = 11.84HH500 pKa = 5.97PPPSKK505 pKa = 10.29RR506 pKa = 11.84KK507 pKa = 7.81RR508 pKa = 11.84QRR510 pKa = 11.84PSSEE514 pKa = 3.49EE515 pKa = 4.05RR516 pKa = 11.84EE517 pKa = 4.11TDD519 pKa = 3.43GSKK522 pKa = 10.91EE523 pKa = 4.45DD524 pKa = 3.53YY525 pKa = 9.18TQASEE530 pKa = 4.55TEE532 pKa = 4.46TEE534 pKa = 4.32TEE536 pKa = 4.13TEE538 pKa = 4.11DD539 pKa = 3.71EE540 pKa = 4.28LGHH543 pKa = 5.65RR544 pKa = 11.84RR545 pKa = 11.84KK546 pKa = 10.11RR547 pKa = 11.84NKK549 pKa = 10.08LRR551 pKa = 11.84ILLDD555 pKa = 3.4RR556 pKa = 11.84LEE558 pKa = 4.37IRR560 pKa = 11.84PHH562 pKa = 6.08ILTNRR567 pKa = 11.84SGNWEE572 pKa = 4.05SASII576 pKa = 3.8
Molecular weight: 68.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
651 |
75 |
576 |
325.5 |
38.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.53 ± 1.118 | 0.922 ± 0.79 |
5.376 ± 0.019 | 5.837 ± 0.979 |
3.226 ± 0.954 | 7.68 ± 3.161 |
3.072 ± 0.42 | 4.455 ± 1.412 |
4.762 ± 1.551 | 7.22 ± 0.956 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.69 ± 0.442 | 3.994 ± 1.204 |
6.298 ± 1.04 | 3.84 ± 1.279 |
10.445 ± 2.313 | 4.762 ± 0.345 |
8.141 ± 1.143 | 3.84 ± 0.072 |
4.147 ± 1.273 | 4.762 ± 2.155 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
