
Sulfitobacter sp. CB2047
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Sulfitobacter; unclassified Sulfitobacter
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
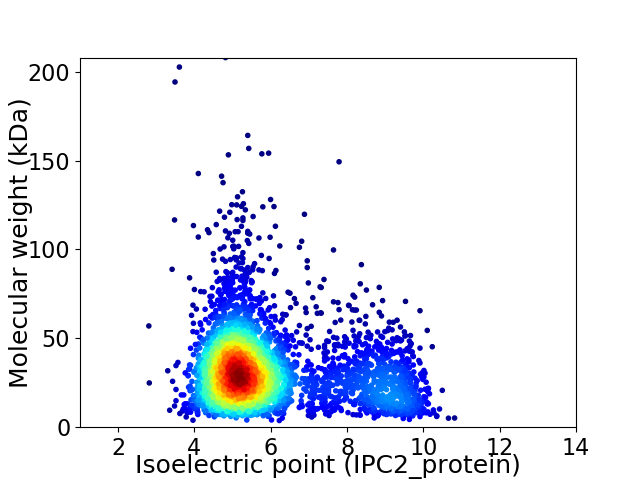
Virtual 2D-PAGE plot for 3563 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
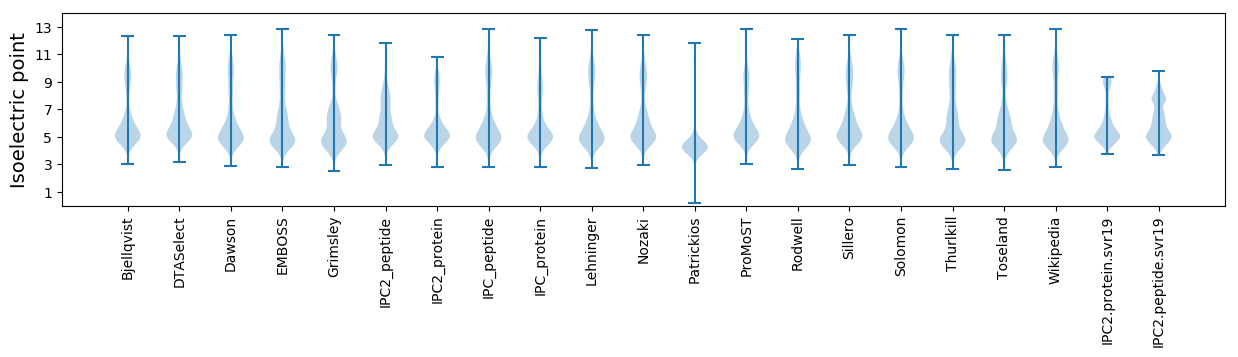
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A085BTE8|A0A085BTE8_9RHOB Phasin PhaP OS=Sulfitobacter sp. CB2047 OX=1525218 GN=IV89_10805 PE=4 SV=1
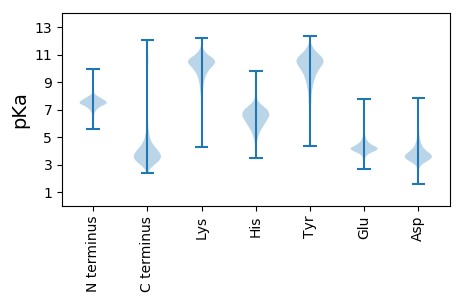
MM1 pKa = 7.7KK2 pKa = 10.34FGTSALALCAFLAACGSGTPFNGEE26 pKa = 4.01TEE28 pKa = 4.35ADD30 pKa = 3.47TGTGTDD36 pKa = 3.35GSTGASAIPEE46 pKa = 4.06EE47 pKa = 4.49LANDD51 pKa = 4.46LKK53 pKa = 11.34SFSYY57 pKa = 10.91DD58 pKa = 3.19PASQTLSITGITADD72 pKa = 4.29DD73 pKa = 4.1SEE75 pKa = 4.52FTANYY80 pKa = 9.3RR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84PGLDD87 pKa = 2.71RR88 pKa = 11.84NGYY91 pKa = 7.93EE92 pKa = 4.96AYY94 pKa = 8.66TAQDD98 pKa = 3.27GSLDD102 pKa = 3.64RR103 pKa = 11.84HH104 pKa = 4.1VTAYY108 pKa = 10.84VKK110 pKa = 10.74DD111 pKa = 3.55IDD113 pKa = 3.85GTRR116 pKa = 11.84AAVVMTGGQFEE127 pKa = 4.61QVFSGAGYY135 pKa = 10.91SNTSYY140 pKa = 10.71SAPVAPGTQSEE151 pKa = 4.78GGLVTYY157 pKa = 10.27AGNYY161 pKa = 9.05IGLLNGAGSGEE172 pKa = 4.25DD173 pKa = 3.68LAPVADD179 pKa = 4.43GTDD182 pKa = 3.8PDD184 pKa = 4.26PLSRR188 pKa = 11.84QAAEE192 pKa = 4.03VTGKK196 pKa = 10.26VVLTGDD202 pKa = 3.5FTDD205 pKa = 3.84AAVAGIIYY213 pKa = 9.63DD214 pKa = 3.77RR215 pKa = 11.84KK216 pKa = 9.55VTDD219 pKa = 3.87FDD221 pKa = 3.55TGGTYY226 pKa = 10.57DD227 pKa = 4.56PNSATPFEE235 pKa = 4.49AQNIALDD242 pKa = 3.8STGIADD248 pKa = 4.29DD249 pKa = 4.48GSFFGTASQSNNAVGEE265 pKa = 4.05YY266 pKa = 10.7GGIFGGTGATEE277 pKa = 4.06VAGVIHH283 pKa = 7.04AEE285 pKa = 3.69NHH287 pKa = 5.16IALGGSGTPIEE298 pKa = 4.46YY299 pKa = 10.26GAFVLTQCGQPGEE312 pKa = 4.93DD313 pKa = 4.64AICDD317 pKa = 3.56QPEE320 pKa = 3.65RR321 pKa = 4.13
MM1 pKa = 7.7KK2 pKa = 10.34FGTSALALCAFLAACGSGTPFNGEE26 pKa = 4.01TEE28 pKa = 4.35ADD30 pKa = 3.47TGTGTDD36 pKa = 3.35GSTGASAIPEE46 pKa = 4.06EE47 pKa = 4.49LANDD51 pKa = 4.46LKK53 pKa = 11.34SFSYY57 pKa = 10.91DD58 pKa = 3.19PASQTLSITGITADD72 pKa = 4.29DD73 pKa = 4.1SEE75 pKa = 4.52FTANYY80 pKa = 9.3RR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84PGLDD87 pKa = 2.71RR88 pKa = 11.84NGYY91 pKa = 7.93EE92 pKa = 4.96AYY94 pKa = 8.66TAQDD98 pKa = 3.27GSLDD102 pKa = 3.64RR103 pKa = 11.84HH104 pKa = 4.1VTAYY108 pKa = 10.84VKK110 pKa = 10.74DD111 pKa = 3.55IDD113 pKa = 3.85GTRR116 pKa = 11.84AAVVMTGGQFEE127 pKa = 4.61QVFSGAGYY135 pKa = 10.91SNTSYY140 pKa = 10.71SAPVAPGTQSEE151 pKa = 4.78GGLVTYY157 pKa = 10.27AGNYY161 pKa = 9.05IGLLNGAGSGEE172 pKa = 4.25DD173 pKa = 3.68LAPVADD179 pKa = 4.43GTDD182 pKa = 3.8PDD184 pKa = 4.26PLSRR188 pKa = 11.84QAAEE192 pKa = 4.03VTGKK196 pKa = 10.26VVLTGDD202 pKa = 3.5FTDD205 pKa = 3.84AAVAGIIYY213 pKa = 9.63DD214 pKa = 3.77RR215 pKa = 11.84KK216 pKa = 9.55VTDD219 pKa = 3.87FDD221 pKa = 3.55TGGTYY226 pKa = 10.57DD227 pKa = 4.56PNSATPFEE235 pKa = 4.49AQNIALDD242 pKa = 3.8STGIADD248 pKa = 4.29DD249 pKa = 4.48GSFFGTASQSNNAVGEE265 pKa = 4.05YY266 pKa = 10.7GGIFGGTGATEE277 pKa = 4.06VAGVIHH283 pKa = 7.04AEE285 pKa = 3.69NHH287 pKa = 5.16IALGGSGTPIEE298 pKa = 4.46YY299 pKa = 10.26GAFVLTQCGQPGEE312 pKa = 4.93DD313 pKa = 4.64AICDD317 pKa = 3.56QPEE320 pKa = 3.65RR321 pKa = 4.13
Molecular weight: 32.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A085BTK0|A0A085BTK0_9RHOB Homoserine dehydrogenase OS=Sulfitobacter sp. CB2047 OX=1525218 GN=IV89_11085 PE=3 SV=1
MM1 pKa = 6.96TAIDD5 pKa = 3.97RR6 pKa = 11.84SVMRR10 pKa = 11.84RR11 pKa = 11.84PQWHH15 pKa = 5.36WWVIIPPVLLAALLFGAAIGEE36 pKa = 4.31VSIPPLVVLKK46 pKa = 10.74VLANKK51 pKa = 9.76ALNAGYY57 pKa = 10.3AVDD60 pKa = 5.53KK61 pKa = 10.47IDD63 pKa = 5.64EE64 pKa = 5.12GIVWNYY70 pKa = 9.69RR71 pKa = 11.84MARR74 pKa = 11.84AVVAMCCGAGLALSGVVLQALLRR97 pKa = 11.84NALADD102 pKa = 4.15PYY104 pKa = 11.24LLGISAGASTGAVAVTILGLGGGAISLSIGAFSGALVAFAFVALLAQAAGGGVGLRR160 pKa = 11.84AAGVIVLAGIAGSQMFNALTALMITKK186 pKa = 9.93SANAEE191 pKa = 3.69QARR194 pKa = 11.84AIMFWLLGNMSGVRR208 pKa = 11.84WPDD211 pKa = 3.23VLSALPAAMIGLLACLWHH229 pKa = 6.88ARR231 pKa = 11.84ALDD234 pKa = 3.66AFTFGSDD241 pKa = 2.95SAASLGIPVRR251 pKa = 11.84RR252 pKa = 11.84VQLLLIGTAALVTAVMVSVVGAIGFVGLVIPHH284 pKa = 7.24AMRR287 pKa = 11.84IVVGHH292 pKa = 5.89RR293 pKa = 11.84HH294 pKa = 6.58AILVPASALAGAVFLVFADD313 pKa = 3.67VVSRR317 pKa = 11.84VIVPGQVLPIGVVTALVGAPVFALILIGKK346 pKa = 7.08RR347 pKa = 11.84TSPP350 pKa = 3.55
MM1 pKa = 6.96TAIDD5 pKa = 3.97RR6 pKa = 11.84SVMRR10 pKa = 11.84RR11 pKa = 11.84PQWHH15 pKa = 5.36WWVIIPPVLLAALLFGAAIGEE36 pKa = 4.31VSIPPLVVLKK46 pKa = 10.74VLANKK51 pKa = 9.76ALNAGYY57 pKa = 10.3AVDD60 pKa = 5.53KK61 pKa = 10.47IDD63 pKa = 5.64EE64 pKa = 5.12GIVWNYY70 pKa = 9.69RR71 pKa = 11.84MARR74 pKa = 11.84AVVAMCCGAGLALSGVVLQALLRR97 pKa = 11.84NALADD102 pKa = 4.15PYY104 pKa = 11.24LLGISAGASTGAVAVTILGLGGGAISLSIGAFSGALVAFAFVALLAQAAGGGVGLRR160 pKa = 11.84AAGVIVLAGIAGSQMFNALTALMITKK186 pKa = 9.93SANAEE191 pKa = 3.69QARR194 pKa = 11.84AIMFWLLGNMSGVRR208 pKa = 11.84WPDD211 pKa = 3.23VLSALPAAMIGLLACLWHH229 pKa = 6.88ARR231 pKa = 11.84ALDD234 pKa = 3.66AFTFGSDD241 pKa = 2.95SAASLGIPVRR251 pKa = 11.84RR252 pKa = 11.84VQLLLIGTAALVTAVMVSVVGAIGFVGLVIPHH284 pKa = 7.24AMRR287 pKa = 11.84IVVGHH292 pKa = 5.89RR293 pKa = 11.84HH294 pKa = 6.58AILVPASALAGAVFLVFADD313 pKa = 3.67VVSRR317 pKa = 11.84VIVPGQVLPIGVVTALVGAPVFALILIGKK346 pKa = 7.08RR347 pKa = 11.84TSPP350 pKa = 3.55
Molecular weight: 35.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1115998 |
32 |
1955 |
313.2 |
33.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.182 ± 0.049 | 0.862 ± 0.015 |
6.335 ± 0.035 | 5.579 ± 0.038 |
3.722 ± 0.026 | 8.568 ± 0.045 |
2.01 ± 0.022 | 5.315 ± 0.031 |
3.471 ± 0.031 | 9.746 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.934 ± 0.023 | 2.776 ± 0.021 |
4.913 ± 0.024 | 3.451 ± 0.025 |
6.186 ± 0.041 | 5.245 ± 0.028 |
5.764 ± 0.029 | 7.337 ± 0.034 |
1.325 ± 0.018 | 2.279 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
