
Alphaproteobacteria bacterium PA2
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; unclassified Alphaproteobacteria
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
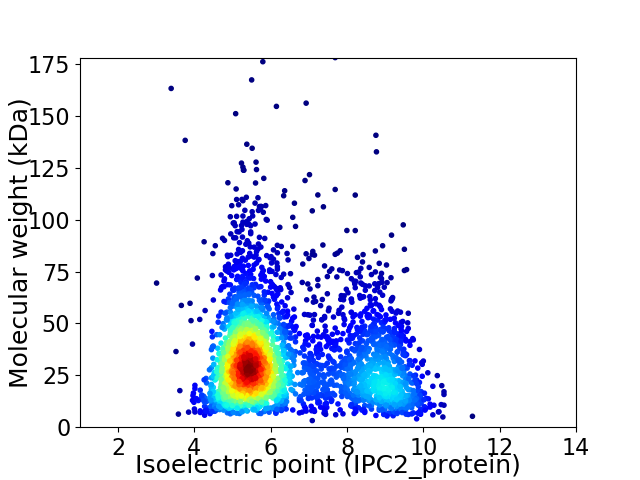
Virtual 2D-PAGE plot for 3257 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
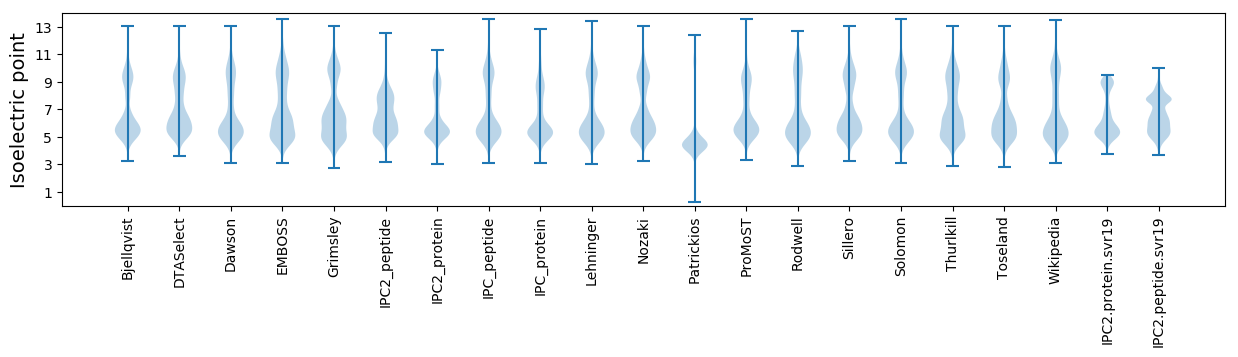
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A257J804|A0A257J804_9PROT Ni_hydr_CYTB domain-containing protein OS=Alphaproteobacteria bacterium PA2 OX=2015570 GN=CFE28_15275 PE=4 SV=1
MM1 pKa = 7.3SWDD4 pKa = 3.37SANAYY9 pKa = 9.85ASSLGGHH16 pKa = 6.9LAFITSAAEE25 pKa = 3.59NDD27 pKa = 3.95YY28 pKa = 10.73LHH30 pKa = 7.21AMAQSDD36 pKa = 4.3VQLLATAPTASDD48 pKa = 3.33GGGARR53 pKa = 11.84YY54 pKa = 8.79LWLGGSDD61 pKa = 3.48AAVEE65 pKa = 4.52GAWRR69 pKa = 11.84WSDD72 pKa = 3.13GSLLSGYY79 pKa = 6.78TNWGAGSLGVEE90 pKa = 3.88PDD92 pKa = 3.46NFFGLQDD99 pKa = 3.52GMALGLQSWPQPSGGIGIAGKK120 pKa = 9.7WNDD123 pKa = 4.4LNPADD128 pKa = 4.64LLYY131 pKa = 10.55FYY133 pKa = 11.22VEE135 pKa = 4.08YY136 pKa = 10.89DD137 pKa = 3.85SIQGTSGNDD146 pKa = 3.17SLIYY150 pKa = 10.01SSSSITSFGGAISIFGGTGNDD171 pKa = 3.49TIEE174 pKa = 4.41AQGGTNYY181 pKa = 10.25LRR183 pKa = 11.84GEE185 pKa = 4.75DD186 pKa = 4.31GNDD189 pKa = 3.77CIIGGTGFDD198 pKa = 5.2DD199 pKa = 3.15IHH201 pKa = 7.16GNKK204 pKa = 9.93GDD206 pKa = 4.69DD207 pKa = 4.01IIIGGPGGDD216 pKa = 3.41WLVGGQGNDD225 pKa = 4.14LITSTGGVDD234 pKa = 3.01ILYY237 pKa = 10.91GNLGNDD243 pKa = 3.97TLNGGPDD250 pKa = 3.76ADD252 pKa = 4.6LMRR255 pKa = 11.84GGQADD260 pKa = 3.91DD261 pKa = 4.37LLNGGGGNDD270 pKa = 3.44WLSGDD275 pKa = 4.32RR276 pKa = 11.84GSDD279 pKa = 3.51TITGGTGADD288 pKa = 3.06IFHH291 pKa = 6.96TFGDD295 pKa = 3.57AGIDD299 pKa = 3.14RR300 pKa = 11.84VTDD303 pKa = 4.08FNLSEE308 pKa = 4.23GDD310 pKa = 3.85RR311 pKa = 11.84VQLDD315 pKa = 3.53PGTVYY320 pKa = 10.56TVTQVGADD328 pKa = 3.41TVINMTGGGQMTLVGVQMATLPAGWIFGAA357 pKa = 4.54
MM1 pKa = 7.3SWDD4 pKa = 3.37SANAYY9 pKa = 9.85ASSLGGHH16 pKa = 6.9LAFITSAAEE25 pKa = 3.59NDD27 pKa = 3.95YY28 pKa = 10.73LHH30 pKa = 7.21AMAQSDD36 pKa = 4.3VQLLATAPTASDD48 pKa = 3.33GGGARR53 pKa = 11.84YY54 pKa = 8.79LWLGGSDD61 pKa = 3.48AAVEE65 pKa = 4.52GAWRR69 pKa = 11.84WSDD72 pKa = 3.13GSLLSGYY79 pKa = 6.78TNWGAGSLGVEE90 pKa = 3.88PDD92 pKa = 3.46NFFGLQDD99 pKa = 3.52GMALGLQSWPQPSGGIGIAGKK120 pKa = 9.7WNDD123 pKa = 4.4LNPADD128 pKa = 4.64LLYY131 pKa = 10.55FYY133 pKa = 11.22VEE135 pKa = 4.08YY136 pKa = 10.89DD137 pKa = 3.85SIQGTSGNDD146 pKa = 3.17SLIYY150 pKa = 10.01SSSSITSFGGAISIFGGTGNDD171 pKa = 3.49TIEE174 pKa = 4.41AQGGTNYY181 pKa = 10.25LRR183 pKa = 11.84GEE185 pKa = 4.75DD186 pKa = 4.31GNDD189 pKa = 3.77CIIGGTGFDD198 pKa = 5.2DD199 pKa = 3.15IHH201 pKa = 7.16GNKK204 pKa = 9.93GDD206 pKa = 4.69DD207 pKa = 4.01IIIGGPGGDD216 pKa = 3.41WLVGGQGNDD225 pKa = 4.14LITSTGGVDD234 pKa = 3.01ILYY237 pKa = 10.91GNLGNDD243 pKa = 3.97TLNGGPDD250 pKa = 3.76ADD252 pKa = 4.6LMRR255 pKa = 11.84GGQADD260 pKa = 3.91DD261 pKa = 4.37LLNGGGGNDD270 pKa = 3.44WLSGDD275 pKa = 4.32RR276 pKa = 11.84GSDD279 pKa = 3.51TITGGTGADD288 pKa = 3.06IFHH291 pKa = 6.96TFGDD295 pKa = 3.57AGIDD299 pKa = 3.14RR300 pKa = 11.84VTDD303 pKa = 4.08FNLSEE308 pKa = 4.23GDD310 pKa = 3.85RR311 pKa = 11.84VQLDD315 pKa = 3.53PGTVYY320 pKa = 10.56TVTQVGADD328 pKa = 3.41TVINMTGGGQMTLVGVQMATLPAGWIFGAA357 pKa = 4.54
Molecular weight: 36.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A257J3X0|A0A257J3X0_9PROT N-acetylmuramoyl-L-alanine amidase OS=Alphaproteobacteria bacterium PA2 OX=2015570 GN=CFE28_06990 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 3.78GWRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.45NGQKK29 pKa = 10.22VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.88GRR39 pKa = 11.84KK40 pKa = 9.04RR41 pKa = 11.84LTAA44 pKa = 4.18
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 3.78GWRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.45NGQKK29 pKa = 10.22VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.88GRR39 pKa = 11.84KK40 pKa = 9.04RR41 pKa = 11.84LTAA44 pKa = 4.18
Molecular weight: 5.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1055213 |
29 |
1685 |
324.0 |
34.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.998 ± 0.052 | 0.772 ± 0.012 |
5.823 ± 0.033 | 5.357 ± 0.039 |
3.672 ± 0.025 | 9.003 ± 0.05 |
1.865 ± 0.019 | 4.711 ± 0.027 |
3.376 ± 0.029 | 10.003 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.474 ± 0.019 | 2.559 ± 0.026 |
5.456 ± 0.035 | 3.268 ± 0.025 |
6.857 ± 0.043 | 5.503 ± 0.032 |
5.34 ± 0.039 | 7.341 ± 0.03 |
1.414 ± 0.019 | 2.207 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
