
Linderina pennispora
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Zoopagomycota; Kickxellomycotina; Kickxellomycetes; Kickxellales; Kickxellaceae; Linderina
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
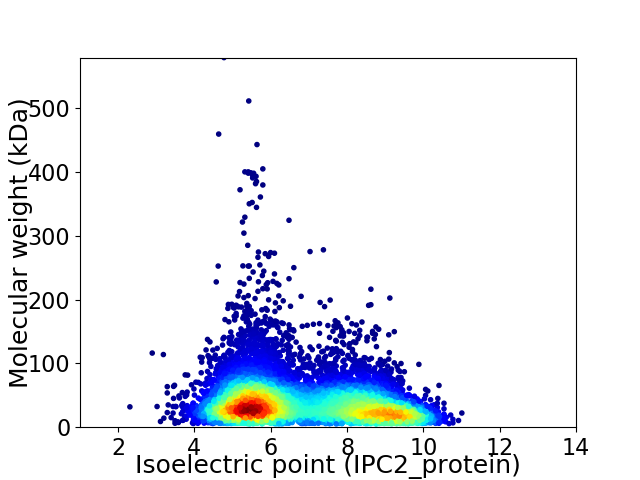
Virtual 2D-PAGE plot for 9274 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1Y1VXJ1|A0A1Y1VXJ1_9FUNG Uncharacterized protein OS=Linderina pennispora OX=61395 GN=DL89DRAFT_270689 PE=4 SV=1
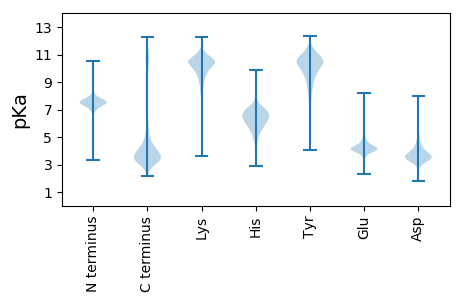
MM1 pKa = 7.18SHH3 pKa = 7.04YY4 pKa = 9.7DD5 pKa = 3.49TAEE8 pKa = 3.92PAADD12 pKa = 4.08DD13 pKa = 4.96DD14 pKa = 4.72FGLVDD19 pKa = 5.72FDD21 pKa = 5.17FADD24 pKa = 3.64SDD26 pKa = 4.17TPAAAHH32 pKa = 6.55AEE34 pKa = 4.41HH35 pKa = 6.78NEE37 pKa = 4.08SPALPGDD44 pKa = 3.64EE45 pKa = 4.1FDD47 pKa = 4.16YY48 pKa = 11.65ANMDD52 pKa = 3.88DD53 pKa = 5.56ADD55 pKa = 4.41FEE57 pKa = 4.64MLVDD61 pKa = 3.98NPPTEE66 pKa = 4.16TATAIHH72 pKa = 7.08SDD74 pKa = 3.69TEE76 pKa = 4.26GAGQHH81 pKa = 5.29TEE83 pKa = 3.86EE84 pKa = 4.69LVDD87 pKa = 4.11FTEE90 pKa = 4.6EE91 pKa = 4.17SLIEE95 pKa = 4.32PDD97 pKa = 5.66SIAEE101 pKa = 4.12DD102 pKa = 3.82AGSSSLPDD110 pKa = 3.63DD111 pKa = 4.57EE112 pKa = 5.36KK113 pKa = 11.58LHH115 pKa = 7.63DD116 pKa = 4.39SLTVNPAEE124 pKa = 4.18SAEE127 pKa = 4.15VSVLAPAPSGHH138 pKa = 7.01ADD140 pKa = 3.48EE141 pKa = 5.31DD142 pKa = 4.65TNGADD147 pKa = 4.54LPAGTPAEE155 pKa = 4.89LIRR158 pKa = 11.84SPALRR163 pKa = 11.84TAGTDD168 pKa = 3.37DD169 pKa = 4.45SLAATDD175 pKa = 4.66NYY177 pKa = 11.18DD178 pKa = 4.35SLVTDD183 pKa = 4.37PFEE186 pKa = 4.76SATSPALPPADD197 pKa = 3.41FVHH200 pKa = 7.07DD201 pKa = 3.89VEE203 pKa = 4.75GTHH206 pKa = 7.03AVDD209 pKa = 3.52HH210 pKa = 6.35SVYY213 pKa = 10.99DD214 pKa = 3.61HH215 pKa = 5.53QTEE218 pKa = 4.07DD219 pKa = 3.62DD220 pKa = 3.81ALVNYY225 pKa = 9.1EE226 pKa = 4.36DD227 pKa = 4.42SDD229 pKa = 4.22VEE231 pKa = 4.4AEE233 pKa = 3.89IEE235 pKa = 4.39EE236 pKa = 4.39IRR238 pKa = 11.84TTDD241 pKa = 3.09PTRR244 pKa = 11.84RR245 pKa = 11.84VPDD248 pKa = 3.46AQVPVTWVFCEE259 pKa = 4.14GDD261 pKa = 3.07WMVYY265 pKa = 10.34LGPEE269 pKa = 3.83QTSYY273 pKa = 11.12DD274 pKa = 3.35AEE276 pKa = 4.23YY277 pKa = 10.67QHH279 pKa = 7.05VLFHH283 pKa = 6.06MSIAQLINNLHH294 pKa = 6.55TDD296 pKa = 3.57TLLKK300 pKa = 10.98DD301 pKa = 3.71EE302 pKa = 4.75VEE304 pKa = 4.3IALEE308 pKa = 4.43FPSLGLKK315 pKa = 9.43IDD317 pKa = 3.6MRR319 pKa = 11.84DD320 pKa = 3.28QEE322 pKa = 4.52SFEE325 pKa = 3.86ITLAQLYY332 pKa = 9.94KK333 pKa = 10.58EE334 pKa = 4.29SQLPSGLASSPYY346 pKa = 8.4FTSASASNYY355 pKa = 9.23CPSPEE360 pKa = 3.81SFAFVIHH367 pKa = 6.13TRR369 pKa = 11.84NSVQSTLEE377 pKa = 4.39HH378 pKa = 6.33IMHH381 pKa = 7.15TLSQHH386 pKa = 5.07EE387 pKa = 4.15ASIEE391 pKa = 3.91AVIPPSEE398 pKa = 4.52DD399 pKa = 3.11PVTVEE404 pKa = 4.34GSVEE408 pKa = 3.97PEE410 pKa = 4.08SVDD413 pKa = 3.57EE414 pKa = 4.82AEE416 pKa = 4.25PAALSEE422 pKa = 4.39EE423 pKa = 4.38AVEE426 pKa = 4.14HH427 pKa = 5.86VPRR430 pKa = 11.84EE431 pKa = 4.08SDD433 pKa = 4.7LEE435 pKa = 4.09DD436 pKa = 3.65HH437 pKa = 6.86SADD440 pKa = 3.59EE441 pKa = 4.38SLEE444 pKa = 4.07AAAA447 pKa = 5.77
MM1 pKa = 7.18SHH3 pKa = 7.04YY4 pKa = 9.7DD5 pKa = 3.49TAEE8 pKa = 3.92PAADD12 pKa = 4.08DD13 pKa = 4.96DD14 pKa = 4.72FGLVDD19 pKa = 5.72FDD21 pKa = 5.17FADD24 pKa = 3.64SDD26 pKa = 4.17TPAAAHH32 pKa = 6.55AEE34 pKa = 4.41HH35 pKa = 6.78NEE37 pKa = 4.08SPALPGDD44 pKa = 3.64EE45 pKa = 4.1FDD47 pKa = 4.16YY48 pKa = 11.65ANMDD52 pKa = 3.88DD53 pKa = 5.56ADD55 pKa = 4.41FEE57 pKa = 4.64MLVDD61 pKa = 3.98NPPTEE66 pKa = 4.16TATAIHH72 pKa = 7.08SDD74 pKa = 3.69TEE76 pKa = 4.26GAGQHH81 pKa = 5.29TEE83 pKa = 3.86EE84 pKa = 4.69LVDD87 pKa = 4.11FTEE90 pKa = 4.6EE91 pKa = 4.17SLIEE95 pKa = 4.32PDD97 pKa = 5.66SIAEE101 pKa = 4.12DD102 pKa = 3.82AGSSSLPDD110 pKa = 3.63DD111 pKa = 4.57EE112 pKa = 5.36KK113 pKa = 11.58LHH115 pKa = 7.63DD116 pKa = 4.39SLTVNPAEE124 pKa = 4.18SAEE127 pKa = 4.15VSVLAPAPSGHH138 pKa = 7.01ADD140 pKa = 3.48EE141 pKa = 5.31DD142 pKa = 4.65TNGADD147 pKa = 4.54LPAGTPAEE155 pKa = 4.89LIRR158 pKa = 11.84SPALRR163 pKa = 11.84TAGTDD168 pKa = 3.37DD169 pKa = 4.45SLAATDD175 pKa = 4.66NYY177 pKa = 11.18DD178 pKa = 4.35SLVTDD183 pKa = 4.37PFEE186 pKa = 4.76SATSPALPPADD197 pKa = 3.41FVHH200 pKa = 7.07DD201 pKa = 3.89VEE203 pKa = 4.75GTHH206 pKa = 7.03AVDD209 pKa = 3.52HH210 pKa = 6.35SVYY213 pKa = 10.99DD214 pKa = 3.61HH215 pKa = 5.53QTEE218 pKa = 4.07DD219 pKa = 3.62DD220 pKa = 3.81ALVNYY225 pKa = 9.1EE226 pKa = 4.36DD227 pKa = 4.42SDD229 pKa = 4.22VEE231 pKa = 4.4AEE233 pKa = 3.89IEE235 pKa = 4.39EE236 pKa = 4.39IRR238 pKa = 11.84TTDD241 pKa = 3.09PTRR244 pKa = 11.84RR245 pKa = 11.84VPDD248 pKa = 3.46AQVPVTWVFCEE259 pKa = 4.14GDD261 pKa = 3.07WMVYY265 pKa = 10.34LGPEE269 pKa = 3.83QTSYY273 pKa = 11.12DD274 pKa = 3.35AEE276 pKa = 4.23YY277 pKa = 10.67QHH279 pKa = 7.05VLFHH283 pKa = 6.06MSIAQLINNLHH294 pKa = 6.55TDD296 pKa = 3.57TLLKK300 pKa = 10.98DD301 pKa = 3.71EE302 pKa = 4.75VEE304 pKa = 4.3IALEE308 pKa = 4.43FPSLGLKK315 pKa = 9.43IDD317 pKa = 3.6MRR319 pKa = 11.84DD320 pKa = 3.28QEE322 pKa = 4.52SFEE325 pKa = 3.86ITLAQLYY332 pKa = 9.94KK333 pKa = 10.58EE334 pKa = 4.29SQLPSGLASSPYY346 pKa = 8.4FTSASASNYY355 pKa = 9.23CPSPEE360 pKa = 3.81SFAFVIHH367 pKa = 6.13TRR369 pKa = 11.84NSVQSTLEE377 pKa = 4.39HH378 pKa = 6.33IMHH381 pKa = 7.15TLSQHH386 pKa = 5.07EE387 pKa = 4.15ASIEE391 pKa = 3.91AVIPPSEE398 pKa = 4.52DD399 pKa = 3.11PVTVEE404 pKa = 4.34GSVEE408 pKa = 3.97PEE410 pKa = 4.08SVDD413 pKa = 3.57EE414 pKa = 4.82AEE416 pKa = 4.25PAALSEE422 pKa = 4.39EE423 pKa = 4.38AVEE426 pKa = 4.14HH427 pKa = 5.86VPRR430 pKa = 11.84EE431 pKa = 4.08SDD433 pKa = 4.7LEE435 pKa = 4.09DD436 pKa = 3.65HH437 pKa = 6.86SADD440 pKa = 3.59EE441 pKa = 4.38SLEE444 pKa = 4.07AAAA447 pKa = 5.77
Molecular weight: 48.37 kDa
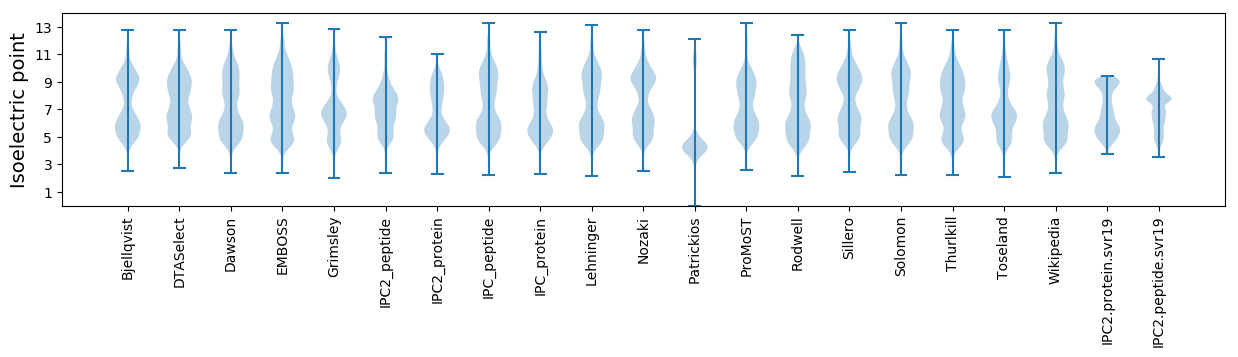
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y1WN60|A0A1Y1WN60_9FUNG Uncharacterized protein OS=Linderina pennispora OX=61395 GN=DL89DRAFT_264784 PE=4 SV=1
MM1 pKa = 7.58LLLPKK6 pKa = 9.34TARR9 pKa = 11.84PSLLLLFTLLLLFTLPPLLMLPLLPTLLQCQFLLRR44 pKa = 11.84PTRR47 pKa = 11.84PRR49 pKa = 11.84VPARR53 pKa = 11.84TFAMVPGSTTPAAAIPVLAAIMSAAAMSRR82 pKa = 11.84AAATTSAAAMNRR94 pKa = 11.84AAATTSAAATSHH106 pKa = 6.45AAAIMSAAAMSRR118 pKa = 11.84AATTIAAAMNRR129 pKa = 11.84AAATTSAAAMSRR141 pKa = 11.84AATTTAAATTTAAAMILAAITIAAATTTAAATMTAAVRR179 pKa = 11.84LATTSSALLSTRR191 pKa = 11.84TAASSTSGATRR202 pKa = 11.84AATATFVPKK211 pKa = 10.6LPQLLIRR218 pKa = 11.84LIPINN223 pKa = 3.74
MM1 pKa = 7.58LLLPKK6 pKa = 9.34TARR9 pKa = 11.84PSLLLLFTLLLLFTLPPLLMLPLLPTLLQCQFLLRR44 pKa = 11.84PTRR47 pKa = 11.84PRR49 pKa = 11.84VPARR53 pKa = 11.84TFAMVPGSTTPAAAIPVLAAIMSAAAMSRR82 pKa = 11.84AAATTSAAAMNRR94 pKa = 11.84AAATTSAAATSHH106 pKa = 6.45AAAIMSAAAMSRR118 pKa = 11.84AATTIAAAMNRR129 pKa = 11.84AAATTSAAAMSRR141 pKa = 11.84AATTTAAATTTAAAMILAAITIAAATTTAAATMTAAVRR179 pKa = 11.84LATTSSALLSTRR191 pKa = 11.84TAASSTSGATRR202 pKa = 11.84AATATFVPKK211 pKa = 10.6LPQLLIRR218 pKa = 11.84LIPINN223 pKa = 3.74
Molecular weight: 22.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3585573 |
49 |
5398 |
386.6 |
42.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.522 ± 0.035 | 1.512 ± 0.012 |
5.576 ± 0.021 | 5.543 ± 0.027 |
3.666 ± 0.017 | 6.552 ± 0.023 |
2.529 ± 0.012 | 4.883 ± 0.021 |
4.409 ± 0.024 | 9.056 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.603 ± 0.012 | 3.455 ± 0.018 |
5.437 ± 0.025 | 4.243 ± 0.022 |
6.221 ± 0.024 | 8.023 ± 0.031 |
5.797 ± 0.023 | 6.784 ± 0.022 |
1.284 ± 0.01 | 2.906 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
