
Alkalinema sp. CACIAM 70d
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Leptolyngbyaceae; Alkalinema; unclassified Alkalinema
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
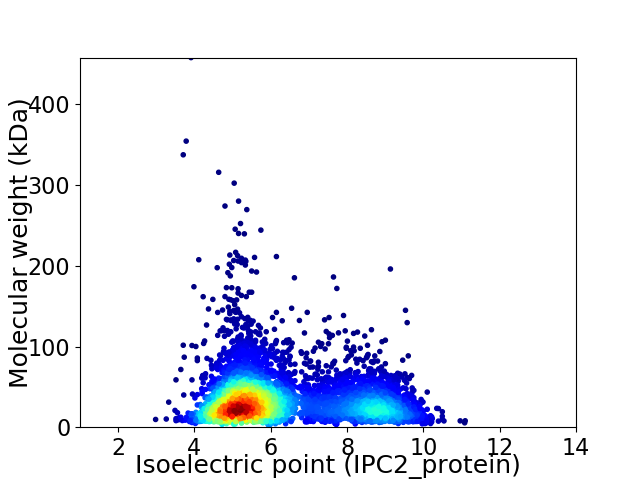
Virtual 2D-PAGE plot for 5211 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A251WKG9|A0A251WKG9_9CYAN AAA_16 domain-containing protein OS=Alkalinema sp. CACIAM 70d OX=1934309 GN=B0A82_02705 PE=4 SV=1
MM1 pKa = 8.03WDD3 pKa = 3.58PSTNTNITLSGSFTIPAGQVTIRR26 pKa = 11.84AAFAPNGKK34 pKa = 10.19LYY36 pKa = 10.89VATNAGGIYY45 pKa = 9.74EE46 pKa = 4.3VNPTDD51 pKa = 3.92GSQISSMITLKK62 pKa = 11.02DD63 pKa = 3.38SANQPLSLAGNGDD76 pKa = 3.48MAFDD80 pKa = 4.39GNGVLYY86 pKa = 10.6LFNDD90 pKa = 3.6ANLYY94 pKa = 8.31TADD97 pKa = 4.25INTQTAYY104 pKa = 11.06LLSTTAGATFNSVAFGSGGNLLAYY128 pKa = 10.09SSSNNTVFSINTNTGIATSVGSPSLGSGIVGSDD161 pKa = 3.97FGSCAQPTPNLTVLKK176 pKa = 10.17SYY178 pKa = 11.39AKK180 pKa = 10.41VGGSAGGTILPGDD193 pKa = 3.75VVEE196 pKa = 4.38YY197 pKa = 10.07TVTITNTGTVPATNSTFSDD216 pKa = 5.2AIPAGATYY224 pKa = 9.22VTGSTQMNSTAVTDD238 pKa = 4.42GSGGTFPFQTARR250 pKa = 11.84NINSPGLINGVVGVGVGYY268 pKa = 10.33QVTLKK273 pKa = 10.77YY274 pKa = 9.98RR275 pKa = 11.84VSLNAVNPPASIVNQGTVLYY295 pKa = 10.23LGNLVPGVVSDD306 pKa = 4.85DD307 pKa = 4.07PNTVAVNDD315 pKa = 4.16ATTITYY321 pKa = 9.35PVVTLSGNVFNDD333 pKa = 3.12SDD335 pKa = 3.93GSKK338 pKa = 10.45VLNGAEE344 pKa = 4.09AGTNAGGLNAVLVNSNNQVVATTAVATNGTYY375 pKa = 10.44SFSNVAVNANYY386 pKa = 8.65TVQITTANATIGSAPPVVTLPSNWVSTGEE415 pKa = 4.08NLNGTVDD422 pKa = 3.61GTVDD426 pKa = 3.18SKK428 pKa = 11.54LAVAVTTNNVTGVNFGIEE446 pKa = 4.06QLPTAVGGTATSQTNPGGTTTVSVPSTLFTGSTDD480 pKa = 3.34PDD482 pKa = 3.67GTVASYY488 pKa = 10.86KK489 pKa = 9.24ITAFPSNATSITINGTNYY507 pKa = 9.76TSASFPGAGVTVTTAQLAGMVVDD530 pKa = 5.74PIDD533 pKa = 3.61GAVNVGILFKK543 pKa = 10.92AIDD546 pKa = 3.62NAGQEE551 pKa = 4.4SSNTATATLPFTAGNVTVSGTVFDD575 pKa = 5.26DD576 pKa = 3.94ADD578 pKa = 3.8GSKK581 pKa = 10.66LLNGTEE587 pKa = 4.01TGTNGGGLNAVLVNGSNQVVATTAVAANGTYY618 pKa = 9.82TFNSVPANANYY629 pKa = 8.58TVQITTATATVGSAPPAIALPSNWVSTGEE658 pKa = 4.09NLNGTIDD665 pKa = 3.66GTVDD669 pKa = 3.1SKK671 pKa = 11.73LSVSVTTNNITGANFGIEE689 pKa = 4.12QLPTAVGGTATSQTNPGGTTTVSVPSTLFTGSTDD723 pKa = 3.34PDD725 pKa = 3.67GTVASYY731 pKa = 10.89KK732 pKa = 9.23ITAFPTNATSITINGTNYY750 pKa = 9.76TSANFPAGGVTVTTAQLAGMVVDD773 pKa = 5.74PIDD776 pKa = 3.78GAVSVGILFKK786 pKa = 10.96AIDD789 pKa = 3.62NAGQEE794 pKa = 4.4SSNTATATLPFTAVVVNNPNLLLVKK819 pKa = 10.37RR820 pKa = 11.84ITAINGTSINFFDD833 pKa = 6.48DD834 pKa = 3.45DD835 pKa = 3.79TTSAKK840 pKa = 10.26KK841 pKa = 10.48DD842 pKa = 4.01DD843 pKa = 5.04DD844 pKa = 3.86NHH846 pKa = 6.84PYY848 pKa = 9.94WPTPLNANGSLGSTSISTFLSGKK871 pKa = 9.44IDD873 pKa = 3.32AGMVKK878 pKa = 10.28SGDD881 pKa = 3.59DD882 pKa = 3.79LEE884 pKa = 4.31YY885 pKa = 10.55TIYY888 pKa = 10.46FLSAGNVPLKK898 pKa = 10.69NVNFCDD904 pKa = 4.33LVPTNTTFQPNSFGPGQGIQLSLGGSFLNLTNVPDD939 pKa = 3.52SDD941 pKa = 3.46RR942 pKa = 11.84AQFLNPGAKK951 pKa = 10.16LPTYY955 pKa = 10.14CPSASNSTGAVLVNIVNSSFAAPDD979 pKa = 3.49NEE981 pKa = 4.8LPNATAPATPGYY993 pKa = 10.19SYY995 pKa = 11.59GFVRR999 pKa = 11.84FKK1001 pKa = 11.47AKK1003 pKa = 10.67VNN1005 pKa = 3.57
MM1 pKa = 8.03WDD3 pKa = 3.58PSTNTNITLSGSFTIPAGQVTIRR26 pKa = 11.84AAFAPNGKK34 pKa = 10.19LYY36 pKa = 10.89VATNAGGIYY45 pKa = 9.74EE46 pKa = 4.3VNPTDD51 pKa = 3.92GSQISSMITLKK62 pKa = 11.02DD63 pKa = 3.38SANQPLSLAGNGDD76 pKa = 3.48MAFDD80 pKa = 4.39GNGVLYY86 pKa = 10.6LFNDD90 pKa = 3.6ANLYY94 pKa = 8.31TADD97 pKa = 4.25INTQTAYY104 pKa = 11.06LLSTTAGATFNSVAFGSGGNLLAYY128 pKa = 10.09SSSNNTVFSINTNTGIATSVGSPSLGSGIVGSDD161 pKa = 3.97FGSCAQPTPNLTVLKK176 pKa = 10.17SYY178 pKa = 11.39AKK180 pKa = 10.41VGGSAGGTILPGDD193 pKa = 3.75VVEE196 pKa = 4.38YY197 pKa = 10.07TVTITNTGTVPATNSTFSDD216 pKa = 5.2AIPAGATYY224 pKa = 9.22VTGSTQMNSTAVTDD238 pKa = 4.42GSGGTFPFQTARR250 pKa = 11.84NINSPGLINGVVGVGVGYY268 pKa = 10.33QVTLKK273 pKa = 10.77YY274 pKa = 9.98RR275 pKa = 11.84VSLNAVNPPASIVNQGTVLYY295 pKa = 10.23LGNLVPGVVSDD306 pKa = 4.85DD307 pKa = 4.07PNTVAVNDD315 pKa = 4.16ATTITYY321 pKa = 9.35PVVTLSGNVFNDD333 pKa = 3.12SDD335 pKa = 3.93GSKK338 pKa = 10.45VLNGAEE344 pKa = 4.09AGTNAGGLNAVLVNSNNQVVATTAVATNGTYY375 pKa = 10.44SFSNVAVNANYY386 pKa = 8.65TVQITTANATIGSAPPVVTLPSNWVSTGEE415 pKa = 4.08NLNGTVDD422 pKa = 3.61GTVDD426 pKa = 3.18SKK428 pKa = 11.54LAVAVTTNNVTGVNFGIEE446 pKa = 4.06QLPTAVGGTATSQTNPGGTTTVSVPSTLFTGSTDD480 pKa = 3.34PDD482 pKa = 3.67GTVASYY488 pKa = 10.86KK489 pKa = 9.24ITAFPSNATSITINGTNYY507 pKa = 9.76TSASFPGAGVTVTTAQLAGMVVDD530 pKa = 5.74PIDD533 pKa = 3.61GAVNVGILFKK543 pKa = 10.92AIDD546 pKa = 3.62NAGQEE551 pKa = 4.4SSNTATATLPFTAGNVTVSGTVFDD575 pKa = 5.26DD576 pKa = 3.94ADD578 pKa = 3.8GSKK581 pKa = 10.66LLNGTEE587 pKa = 4.01TGTNGGGLNAVLVNGSNQVVATTAVAANGTYY618 pKa = 9.82TFNSVPANANYY629 pKa = 8.58TVQITTATATVGSAPPAIALPSNWVSTGEE658 pKa = 4.09NLNGTIDD665 pKa = 3.66GTVDD669 pKa = 3.1SKK671 pKa = 11.73LSVSVTTNNITGANFGIEE689 pKa = 4.12QLPTAVGGTATSQTNPGGTTTVSVPSTLFTGSTDD723 pKa = 3.34PDD725 pKa = 3.67GTVASYY731 pKa = 10.89KK732 pKa = 9.23ITAFPTNATSITINGTNYY750 pKa = 9.76TSANFPAGGVTVTTAQLAGMVVDD773 pKa = 5.74PIDD776 pKa = 3.78GAVSVGILFKK786 pKa = 10.96AIDD789 pKa = 3.62NAGQEE794 pKa = 4.4SSNTATATLPFTAVVVNNPNLLLVKK819 pKa = 10.37RR820 pKa = 11.84ITAINGTSINFFDD833 pKa = 6.48DD834 pKa = 3.45DD835 pKa = 3.79TTSAKK840 pKa = 10.26KK841 pKa = 10.48DD842 pKa = 4.01DD843 pKa = 5.04DD844 pKa = 3.86NHH846 pKa = 6.84PYY848 pKa = 9.94WPTPLNANGSLGSTSISTFLSGKK871 pKa = 9.44IDD873 pKa = 3.32AGMVKK878 pKa = 10.28SGDD881 pKa = 3.59DD882 pKa = 3.79LEE884 pKa = 4.31YY885 pKa = 10.55TIYY888 pKa = 10.46FLSAGNVPLKK898 pKa = 10.69NVNFCDD904 pKa = 4.33LVPTNTTFQPNSFGPGQGIQLSLGGSFLNLTNVPDD939 pKa = 3.52SDD941 pKa = 3.46RR942 pKa = 11.84AQFLNPGAKK951 pKa = 10.16LPTYY955 pKa = 10.14CPSASNSTGAVLVNIVNSSFAAPDD979 pKa = 3.49NEE981 pKa = 4.8LPNATAPATPGYY993 pKa = 10.19SYY995 pKa = 11.59GFVRR999 pKa = 11.84FKK1001 pKa = 11.47AKK1003 pKa = 10.67VNN1005 pKa = 3.57
Molecular weight: 101.25 kDa
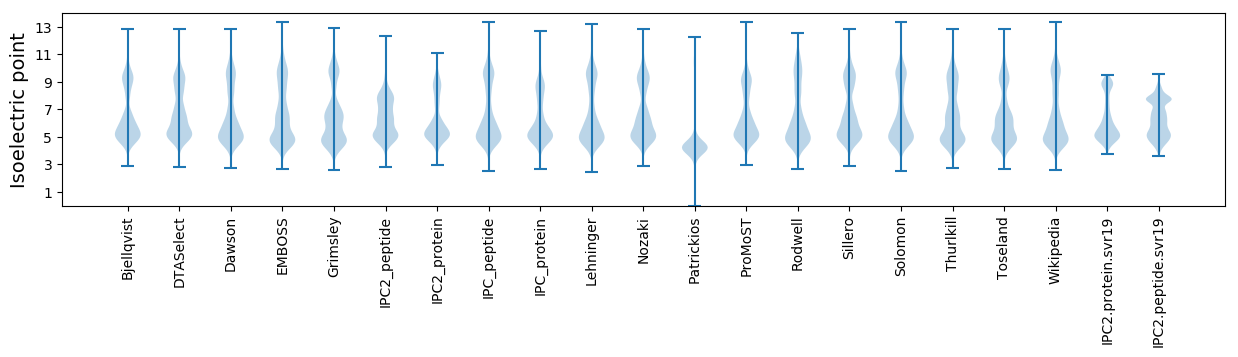
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A251WHR2|A0A251WHR2_9CYAN Uncharacterized protein OS=Alkalinema sp. CACIAM 70d OX=1934309 GN=B0A82_10095 PE=4 SV=1
MM1 pKa = 7.53SLPLPPKK8 pKa = 9.33SQSSPSPPTRR18 pKa = 11.84VSLPRR23 pKa = 11.84LPARR27 pKa = 11.84KK28 pKa = 9.22SLPASPLRR36 pKa = 11.84KK37 pKa = 9.58LLALLPIIALSRR49 pKa = 11.84LLPMPFIAAVPSSRR63 pKa = 11.84FKK65 pKa = 10.77FSRR68 pKa = 11.84LSPRR72 pKa = 11.84VV73 pKa = 3.27
MM1 pKa = 7.53SLPLPPKK8 pKa = 9.33SQSSPSPPTRR18 pKa = 11.84VSLPRR23 pKa = 11.84LPARR27 pKa = 11.84KK28 pKa = 9.22SLPASPLRR36 pKa = 11.84KK37 pKa = 9.58LLALLPIIALSRR49 pKa = 11.84LLPMPFIAAVPSSRR63 pKa = 11.84FKK65 pKa = 10.77FSRR68 pKa = 11.84LSPRR72 pKa = 11.84VV73 pKa = 3.27
Molecular weight: 7.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1707141 |
29 |
4319 |
327.6 |
36.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.013 ± 0.038 | 0.956 ± 0.012 |
5.216 ± 0.029 | 5.729 ± 0.035 |
3.706 ± 0.02 | 6.92 ± 0.038 |
1.951 ± 0.019 | 5.949 ± 0.025 |
3.797 ± 0.026 | 11.087 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.959 ± 0.015 | 3.673 ± 0.03 |
5.246 ± 0.036 | 5.943 ± 0.035 |
5.686 ± 0.031 | 6.319 ± 0.033 |
5.868 ± 0.033 | 6.635 ± 0.026 |
1.531 ± 0.014 | 2.814 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
