
Candidatus Chloroploca asiatica
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Chloroflexia; Chloroflexales; Chloroflexineae; Oscillochloridaceae; Candidatus Chloroploca
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
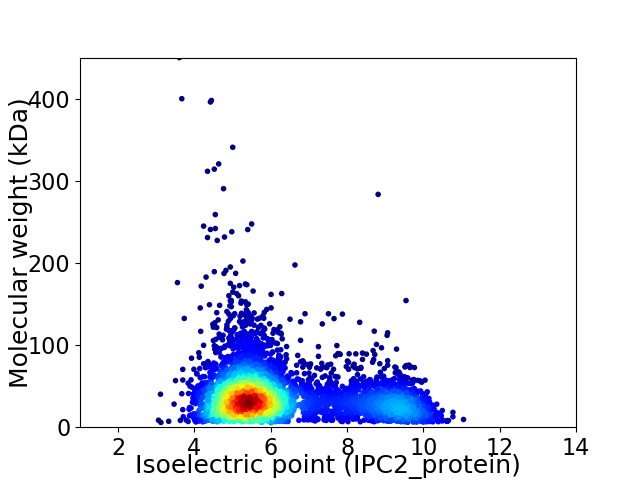
Virtual 2D-PAGE plot for 4456 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H3KNG9|A0A2H3KNG9_9CHLR NADPH quinone oxidoreductase OS=Candidatus Chloroploca asiatica OX=1506545 GN=A9Q02_00100 PE=4 SV=1
MM1 pKa = 7.73KK2 pKa = 10.51LRR4 pKa = 11.84MTILSALLMGMLVLAACGTAPSTGSTTEE32 pKa = 3.85PTPATTEE39 pKa = 3.67QEE41 pKa = 4.43TPVTEE46 pKa = 4.15EE47 pKa = 3.65ATTPEE52 pKa = 4.41ASDD55 pKa = 3.79ALAGTSWVLTQLNGAAPLSEE75 pKa = 4.14GTPISAEE82 pKa = 3.75FDD84 pKa = 3.78TEE86 pKa = 4.33GSVAGSAGCNRR97 pKa = 11.84YY98 pKa = 8.49FAGYY102 pKa = 8.16TIADD106 pKa = 3.81GTLSITQAGSTMMACEE122 pKa = 4.23DD123 pKa = 4.16AIMAQEE129 pKa = 4.26QAFLDD134 pKa = 3.71QFSRR138 pKa = 11.84ATSYY142 pKa = 11.44VIDD145 pKa = 4.4GATLTITIDD154 pKa = 3.68DD155 pKa = 3.9GTTLVFEE162 pKa = 4.45RR163 pKa = 11.84AEE165 pKa = 4.22EE166 pKa = 4.04EE167 pKa = 4.43VTTSNDD173 pKa = 3.03LTGTNWVLTAINGAAPLSEE192 pKa = 4.38SAPISAEE199 pKa = 3.28FDD201 pKa = 3.35TEE203 pKa = 4.07GRR205 pKa = 11.84ITGSAGCNRR214 pKa = 11.84YY215 pKa = 8.65FADD218 pKa = 3.51YY219 pKa = 9.59TVADD223 pKa = 4.1GTLSISQAGSTRR235 pKa = 11.84MACEE239 pKa = 3.9EE240 pKa = 4.46AIMAQEE246 pKa = 4.29QAFLDD251 pKa = 3.73QLSRR255 pKa = 11.84ATSYY259 pKa = 11.43VIDD262 pKa = 4.4GATLTITIDD271 pKa = 3.65DD272 pKa = 4.2GATLVFEE279 pKa = 4.4QTSNALAGTSWLLTEE294 pKa = 5.17LNGAAPLSEE303 pKa = 4.31ATPISAEE310 pKa = 3.75FDD312 pKa = 3.54DD313 pKa = 4.57EE314 pKa = 4.54GRR316 pKa = 11.84VAGSAGCNRR325 pKa = 11.84YY326 pKa = 8.51FAGYY330 pKa = 7.35TVAEE334 pKa = 4.53GTLSITQAGSTMMACEE350 pKa = 4.08DD351 pKa = 4.48AILAQEE357 pKa = 4.17MAFLDD362 pKa = 3.69QLSRR366 pKa = 11.84ATSYY370 pKa = 11.45VIDD373 pKa = 4.12GTTLTITVDD382 pKa = 3.47DD383 pKa = 4.49GATLVFVRR391 pKa = 11.84AA392 pKa = 3.97
MM1 pKa = 7.73KK2 pKa = 10.51LRR4 pKa = 11.84MTILSALLMGMLVLAACGTAPSTGSTTEE32 pKa = 3.85PTPATTEE39 pKa = 3.67QEE41 pKa = 4.43TPVTEE46 pKa = 4.15EE47 pKa = 3.65ATTPEE52 pKa = 4.41ASDD55 pKa = 3.79ALAGTSWVLTQLNGAAPLSEE75 pKa = 4.14GTPISAEE82 pKa = 3.75FDD84 pKa = 3.78TEE86 pKa = 4.33GSVAGSAGCNRR97 pKa = 11.84YY98 pKa = 8.49FAGYY102 pKa = 8.16TIADD106 pKa = 3.81GTLSITQAGSTMMACEE122 pKa = 4.23DD123 pKa = 4.16AIMAQEE129 pKa = 4.26QAFLDD134 pKa = 3.71QFSRR138 pKa = 11.84ATSYY142 pKa = 11.44VIDD145 pKa = 4.4GATLTITIDD154 pKa = 3.68DD155 pKa = 3.9GTTLVFEE162 pKa = 4.45RR163 pKa = 11.84AEE165 pKa = 4.22EE166 pKa = 4.04EE167 pKa = 4.43VTTSNDD173 pKa = 3.03LTGTNWVLTAINGAAPLSEE192 pKa = 4.38SAPISAEE199 pKa = 3.28FDD201 pKa = 3.35TEE203 pKa = 4.07GRR205 pKa = 11.84ITGSAGCNRR214 pKa = 11.84YY215 pKa = 8.65FADD218 pKa = 3.51YY219 pKa = 9.59TVADD223 pKa = 4.1GTLSISQAGSTRR235 pKa = 11.84MACEE239 pKa = 3.9EE240 pKa = 4.46AIMAQEE246 pKa = 4.29QAFLDD251 pKa = 3.73QLSRR255 pKa = 11.84ATSYY259 pKa = 11.43VIDD262 pKa = 4.4GATLTITIDD271 pKa = 3.65DD272 pKa = 4.2GATLVFEE279 pKa = 4.4QTSNALAGTSWLLTEE294 pKa = 5.17LNGAAPLSEE303 pKa = 4.31ATPISAEE310 pKa = 3.75FDD312 pKa = 3.54DD313 pKa = 4.57EE314 pKa = 4.54GRR316 pKa = 11.84VAGSAGCNRR325 pKa = 11.84YY326 pKa = 8.51FAGYY330 pKa = 7.35TVAEE334 pKa = 4.53GTLSITQAGSTMMACEE350 pKa = 4.08DD351 pKa = 4.48AILAQEE357 pKa = 4.17MAFLDD362 pKa = 3.69QLSRR366 pKa = 11.84ATSYY370 pKa = 11.45VIDD373 pKa = 4.12GTTLTITVDD382 pKa = 3.47DD383 pKa = 4.49GATLVFVRR391 pKa = 11.84AA392 pKa = 3.97
Molecular weight: 40.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H3KKB4|A0A2H3KKB4_9CHLR ATPase/protein kinase OS=Candidatus Chloroploca asiatica OX=1506545 GN=A9Q02_15515 PE=3 SV=1
MM1 pKa = 7.38AAAPIGPAGAMPAPANGRR19 pKa = 11.84IPGSDD24 pKa = 2.93RR25 pKa = 11.84SIHH28 pKa = 5.67ARR30 pKa = 11.84AIASSVYY37 pKa = 10.44APRR40 pKa = 11.84QGTRR44 pKa = 11.84KK45 pKa = 8.34GTRR48 pKa = 11.84KK49 pKa = 8.47GTRR52 pKa = 11.84KK53 pKa = 8.47GTRR56 pKa = 11.84KK57 pKa = 8.47GTRR60 pKa = 11.84KK61 pKa = 8.47GTRR64 pKa = 11.84KK65 pKa = 8.47GTRR68 pKa = 11.84KK69 pKa = 8.47GTRR72 pKa = 11.84KK73 pKa = 8.47GTRR76 pKa = 11.84KK77 pKa = 8.47GTRR80 pKa = 11.84KK81 pKa = 8.47GTRR84 pKa = 11.84KK85 pKa = 8.5GTRR88 pKa = 11.84KK89 pKa = 10.05GG90 pKa = 3.32
MM1 pKa = 7.38AAAPIGPAGAMPAPANGRR19 pKa = 11.84IPGSDD24 pKa = 2.93RR25 pKa = 11.84SIHH28 pKa = 5.67ARR30 pKa = 11.84AIASSVYY37 pKa = 10.44APRR40 pKa = 11.84QGTRR44 pKa = 11.84KK45 pKa = 8.34GTRR48 pKa = 11.84KK49 pKa = 8.47GTRR52 pKa = 11.84KK53 pKa = 8.47GTRR56 pKa = 11.84KK57 pKa = 8.47GTRR60 pKa = 11.84KK61 pKa = 8.47GTRR64 pKa = 11.84KK65 pKa = 8.47GTRR68 pKa = 11.84KK69 pKa = 8.47GTRR72 pKa = 11.84KK73 pKa = 8.47GTRR76 pKa = 11.84KK77 pKa = 8.47GTRR80 pKa = 11.84KK81 pKa = 8.47GTRR84 pKa = 11.84KK85 pKa = 8.5GTRR88 pKa = 11.84KK89 pKa = 10.05GG90 pKa = 3.32
Molecular weight: 9.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1658142 |
52 |
4279 |
372.1 |
40.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.531 ± 0.055 | 0.785 ± 0.011 |
5.044 ± 0.03 | 5.672 ± 0.036 |
3.355 ± 0.022 | 7.883 ± 0.04 |
2.223 ± 0.021 | 5.0 ± 0.031 |
1.866 ± 0.026 | 11.816 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.146 ± 0.017 | 2.597 ± 0.031 |
5.814 ± 0.029 | 4.209 ± 0.027 |
7.145 ± 0.037 | 4.975 ± 0.029 |
6.148 ± 0.052 | 7.628 ± 0.031 |
1.513 ± 0.019 | 2.65 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
