
Rhizomicrobium sp. SCGC AG-212-E05
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Micropepsales; Micropepsaceae; Rhizomicrobium; unclassified Rhizomicrobium
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins
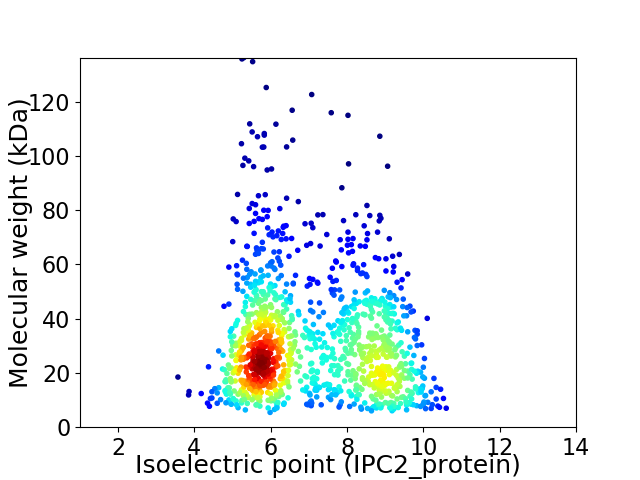
Virtual 2D-PAGE plot for 1220 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A177QAG0|A0A177QAG0_9PROT Uncharacterized protein OS=Rhizomicrobium sp. SCGC AG-212-E05 OX=1799655 GN=AYO42_00935 PE=4 SV=1
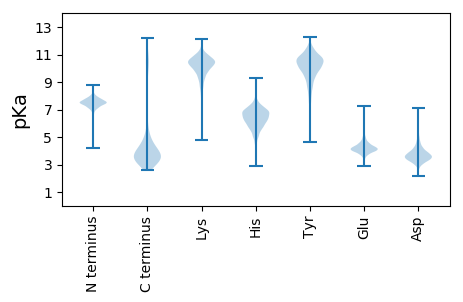
MM1 pKa = 7.74ASLITIGICVYY12 pKa = 10.56DD13 pKa = 4.16YY14 pKa = 7.39YY15 pKa = 11.11TPEE18 pKa = 3.94TGLIGSGGVILVAGAAVLMFLAALVMARR46 pKa = 11.84MPGRR50 pKa = 11.84GSLLQTFLVVSILLDD65 pKa = 3.78IIGSSVAGYY74 pKa = 9.77FLEE77 pKa = 4.72SPVIMAAQALALVGWVINVVSNAAEE102 pKa = 3.73RR103 pKa = 11.84SVEE106 pKa = 3.87TDD108 pKa = 3.09EE109 pKa = 6.38VEE111 pKa = 5.92DD112 pKa = 3.95EE113 pKa = 4.37DD114 pKa = 4.71
MM1 pKa = 7.74ASLITIGICVYY12 pKa = 10.56DD13 pKa = 4.16YY14 pKa = 7.39YY15 pKa = 11.11TPEE18 pKa = 3.94TGLIGSGGVILVAGAAVLMFLAALVMARR46 pKa = 11.84MPGRR50 pKa = 11.84GSLLQTFLVVSILLDD65 pKa = 3.78IIGSSVAGYY74 pKa = 9.77FLEE77 pKa = 4.72SPVIMAAQALALVGWVINVVSNAAEE102 pKa = 3.73RR103 pKa = 11.84SVEE106 pKa = 3.87TDD108 pKa = 3.09EE109 pKa = 6.38VEE111 pKa = 5.92DD112 pKa = 3.95EE113 pKa = 4.37DD114 pKa = 4.71
Molecular weight: 11.9 kDa
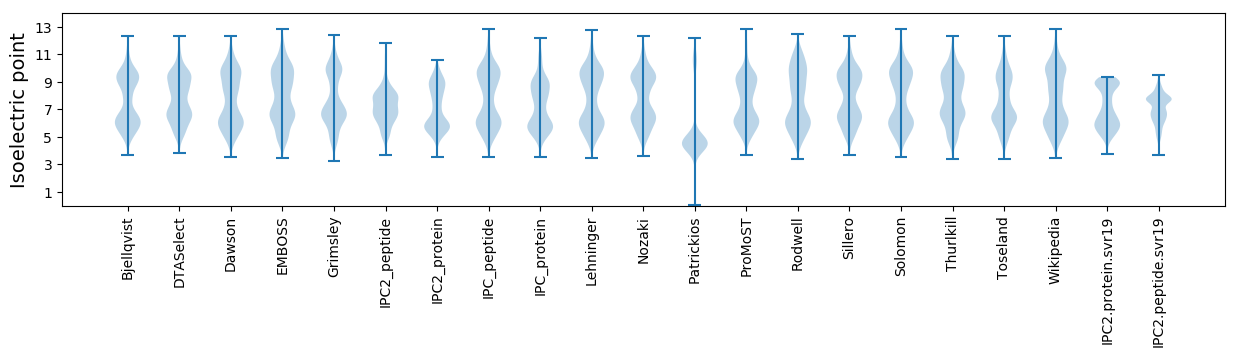
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A177Q4M8|A0A177Q4M8_9PROT Uncharacterized protein OS=Rhizomicrobium sp. SCGC AG-212-E05 OX=1799655 GN=AYO42_06595 PE=4 SV=1
MM1 pKa = 7.65KK2 pKa = 10.62KK3 pKa = 8.94LTLLTALLCLGTAAPALAAWNEE25 pKa = 3.8IGTIRR30 pKa = 11.84VNPGQDD36 pKa = 2.55RR37 pKa = 11.84DD38 pKa = 3.75VRR40 pKa = 11.84NFDD43 pKa = 3.31LGGPVDD49 pKa = 4.15RR50 pKa = 11.84LQLRR54 pKa = 11.84AEE56 pKa = 4.47GADD59 pKa = 3.14IRR61 pKa = 11.84CRR63 pKa = 11.84AITARR68 pKa = 11.84FGNGRR73 pKa = 11.84SQTLYY78 pKa = 10.55QGNLTEE84 pKa = 6.19GMPQNINLPGRR95 pKa = 11.84EE96 pKa = 3.81RR97 pKa = 11.84NITRR101 pKa = 11.84LTFEE105 pKa = 5.07CGAEE109 pKa = 3.69RR110 pKa = 11.84QVGNRR115 pKa = 11.84RR116 pKa = 11.84GDD118 pKa = 3.22RR119 pKa = 11.84DD120 pKa = 3.16DD121 pKa = 3.45RR122 pKa = 11.84RR123 pKa = 11.84RR124 pKa = 11.84DD125 pKa = 3.18RR126 pKa = 11.84RR127 pKa = 11.84GAGDD131 pKa = 3.42ASIVVVADD139 pKa = 2.8VDD141 pKa = 3.94RR142 pKa = 11.84YY143 pKa = 9.49RR144 pKa = 11.84KK145 pKa = 8.14QWRR148 pKa = 11.84SNPRR152 pKa = 11.84YY153 pKa = 8.48QNEE156 pKa = 3.61WSKK159 pKa = 11.04IFNWGSDD166 pKa = 3.35MVNDD170 pKa = 3.47WRR172 pKa = 11.84LLSTVSFEE180 pKa = 3.98GRR182 pKa = 11.84RR183 pKa = 11.84DD184 pKa = 3.43SEE186 pKa = 4.08RR187 pKa = 11.84AFAGWKK193 pKa = 8.36GQNVDD198 pKa = 4.33AVALMPLDD206 pKa = 4.11ADD208 pKa = 3.83VRR210 pKa = 11.84CNRR213 pKa = 11.84VSARR217 pKa = 11.84FGNGQAQPLNVNNGDD232 pKa = 3.63YY233 pKa = 10.75LRR235 pKa = 11.84RR236 pKa = 11.84GQYY239 pKa = 10.55YY240 pKa = 10.72KK241 pKa = 10.98LDD243 pKa = 3.72LPGRR247 pKa = 11.84ARR249 pKa = 11.84DD250 pKa = 3.84LVSLDD255 pKa = 3.2MRR257 pKa = 11.84CRR259 pKa = 11.84ATNSDD264 pKa = 3.44RR265 pKa = 11.84VRR267 pKa = 11.84VQIFTSRR274 pKa = 3.06
MM1 pKa = 7.65KK2 pKa = 10.62KK3 pKa = 8.94LTLLTALLCLGTAAPALAAWNEE25 pKa = 3.8IGTIRR30 pKa = 11.84VNPGQDD36 pKa = 2.55RR37 pKa = 11.84DD38 pKa = 3.75VRR40 pKa = 11.84NFDD43 pKa = 3.31LGGPVDD49 pKa = 4.15RR50 pKa = 11.84LQLRR54 pKa = 11.84AEE56 pKa = 4.47GADD59 pKa = 3.14IRR61 pKa = 11.84CRR63 pKa = 11.84AITARR68 pKa = 11.84FGNGRR73 pKa = 11.84SQTLYY78 pKa = 10.55QGNLTEE84 pKa = 6.19GMPQNINLPGRR95 pKa = 11.84EE96 pKa = 3.81RR97 pKa = 11.84NITRR101 pKa = 11.84LTFEE105 pKa = 5.07CGAEE109 pKa = 3.69RR110 pKa = 11.84QVGNRR115 pKa = 11.84RR116 pKa = 11.84GDD118 pKa = 3.22RR119 pKa = 11.84DD120 pKa = 3.16DD121 pKa = 3.45RR122 pKa = 11.84RR123 pKa = 11.84RR124 pKa = 11.84DD125 pKa = 3.18RR126 pKa = 11.84RR127 pKa = 11.84GAGDD131 pKa = 3.42ASIVVVADD139 pKa = 2.8VDD141 pKa = 3.94RR142 pKa = 11.84YY143 pKa = 9.49RR144 pKa = 11.84KK145 pKa = 8.14QWRR148 pKa = 11.84SNPRR152 pKa = 11.84YY153 pKa = 8.48QNEE156 pKa = 3.61WSKK159 pKa = 11.04IFNWGSDD166 pKa = 3.35MVNDD170 pKa = 3.47WRR172 pKa = 11.84LLSTVSFEE180 pKa = 3.98GRR182 pKa = 11.84RR183 pKa = 11.84DD184 pKa = 3.43SEE186 pKa = 4.08RR187 pKa = 11.84AFAGWKK193 pKa = 8.36GQNVDD198 pKa = 4.33AVALMPLDD206 pKa = 4.11ADD208 pKa = 3.83VRR210 pKa = 11.84CNRR213 pKa = 11.84VSARR217 pKa = 11.84FGNGQAQPLNVNNGDD232 pKa = 3.63YY233 pKa = 10.75LRR235 pKa = 11.84RR236 pKa = 11.84GQYY239 pKa = 10.55YY240 pKa = 10.72KK241 pKa = 10.98LDD243 pKa = 3.72LPGRR247 pKa = 11.84ARR249 pKa = 11.84DD250 pKa = 3.84LVSLDD255 pKa = 3.2MRR257 pKa = 11.84CRR259 pKa = 11.84ATNSDD264 pKa = 3.44RR265 pKa = 11.84VRR267 pKa = 11.84VQIFTSRR274 pKa = 3.06
Molecular weight: 31.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
369753 |
52 |
1237 |
303.1 |
32.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.43 ± 0.084 | 0.856 ± 0.019 |
5.49 ± 0.047 | 5.086 ± 0.058 |
3.842 ± 0.045 | 8.712 ± 0.065 |
2.055 ± 0.037 | 5.147 ± 0.041 |
4.131 ± 0.056 | 9.9 ± 0.072 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.581 ± 0.031 | 2.897 ± 0.043 |
5.242 ± 0.046 | 3.303 ± 0.039 |
6.702 ± 0.063 | 5.397 ± 0.043 |
5.272 ± 0.055 | 7.168 ± 0.052 |
1.352 ± 0.027 | 2.436 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
