
Cattle blood-associated circovirus-like virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; Porprismacovirus bovas2
Average proteome isoelectric point is 8.55
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
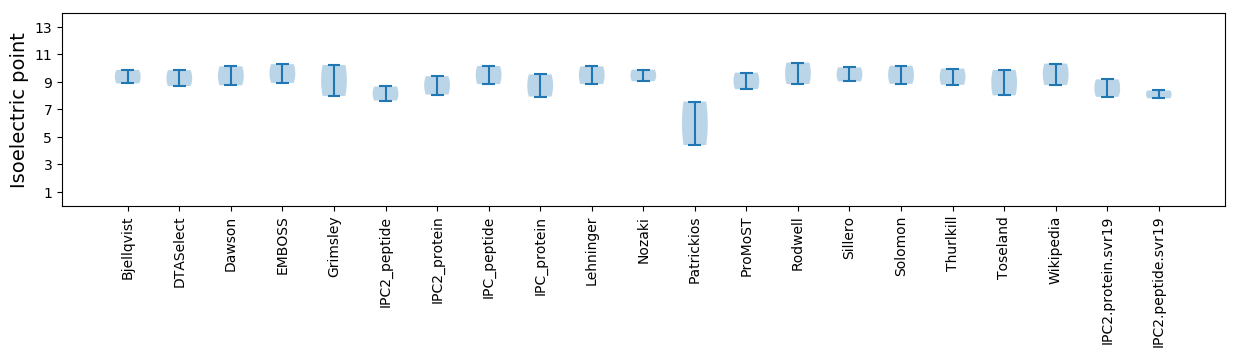
* You can choose from 21 different methods for calculating isoelectric point
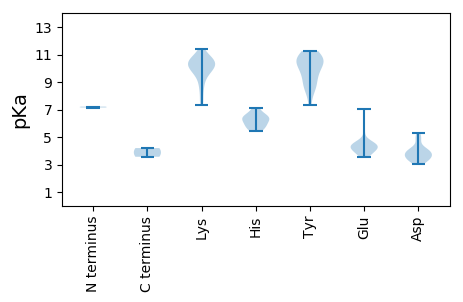
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2L0HH22|A0A2L0HH22_9VIRU Replication-associated protein OS=Cattle blood-associated circovirus-like virus OX=2077298 PE=4 SV=1
MM1 pKa = 7.14SQSQKK6 pKa = 9.97IEE8 pKa = 4.05KK9 pKa = 9.82VFQSQARR16 pKa = 11.84RR17 pKa = 11.84FHH19 pKa = 6.42IVANEE24 pKa = 3.56KK25 pKa = 10.49SLPYY29 pKa = 10.49LNNILNFLTTKK40 pKa = 9.46PYY42 pKa = 11.07VYY44 pKa = 10.01ILCRR48 pKa = 11.84KK49 pKa = 9.54GYY51 pKa = 9.5NKK53 pKa = 10.14RR54 pKa = 11.84NCEE57 pKa = 3.7HH58 pKa = 5.95AHH60 pKa = 6.25IYY62 pKa = 10.76VIFKK66 pKa = 10.33KK67 pKa = 10.42AVRR70 pKa = 11.84LSNKK74 pKa = 7.36NTHH77 pKa = 5.8GCHH80 pKa = 5.69IAKK83 pKa = 10.09CRR85 pKa = 11.84GTTQQNIEE93 pKa = 4.59YY94 pKa = 9.57IKK96 pKa = 9.95GHH98 pKa = 6.53HH99 pKa = 6.29PMEE102 pKa = 4.42VCEE105 pKa = 4.36KK106 pKa = 11.38GEE108 pKa = 3.91IPEE111 pKa = 4.3NDD113 pKa = 3.27GDD115 pKa = 3.88IPPRR119 pKa = 11.84KK120 pKa = 8.56WDD122 pKa = 3.69GFVQQIKK129 pKa = 9.33QKK131 pKa = 10.69KK132 pKa = 7.94IDD134 pKa = 3.59KK135 pKa = 9.12FDD137 pKa = 3.63PMFARR142 pKa = 11.84YY143 pKa = 8.88EE144 pKa = 4.36GYY146 pKa = 10.61ANRR149 pKa = 11.84RR150 pKa = 11.84VAEE153 pKa = 3.95IMVEE157 pKa = 3.94EE158 pKa = 4.54LEE160 pKa = 5.51DD161 pKa = 3.95YY162 pKa = 11.19EE163 pKa = 7.06GDD165 pKa = 4.93LKK167 pKa = 11.35DD168 pKa = 3.89KK169 pKa = 11.04NCWIYY174 pKa = 11.25GEE176 pKa = 4.61PGTGKK181 pKa = 10.19SYY183 pKa = 10.56SARR186 pKa = 11.84YY187 pKa = 8.61CKK189 pKa = 9.98RR190 pKa = 11.84SKK192 pKa = 10.18IYY194 pKa = 9.41PKK196 pKa = 10.14PVSKK200 pKa = 9.54WWDD203 pKa = 3.5GYY205 pKa = 10.74KK206 pKa = 9.9GQKK209 pKa = 8.48VVVMDD214 pKa = 5.31DD215 pKa = 4.23LDD217 pKa = 4.1PDD219 pKa = 4.0RR220 pKa = 11.84AKK222 pKa = 10.46MLADD226 pKa = 4.83KK227 pKa = 10.84IKK229 pKa = 9.91IWTDD233 pKa = 3.0KK234 pKa = 10.55YY235 pKa = 10.86PFVAEE240 pKa = 4.47VKK242 pKa = 10.34SSSAVVDD249 pKa = 3.36PRR251 pKa = 11.84INFIVTSNYY260 pKa = 10.33SIEE263 pKa = 4.36DD264 pKa = 3.64CFEE267 pKa = 4.5SKK269 pKa = 10.57DD270 pKa = 3.4VEE272 pKa = 4.43AIKK275 pKa = 10.7RR276 pKa = 11.84RR277 pKa = 11.84FTVLHH282 pKa = 5.42FTGEE286 pKa = 4.39SIEE289 pKa = 4.25EE290 pKa = 4.06EE291 pKa = 4.2
MM1 pKa = 7.14SQSQKK6 pKa = 9.97IEE8 pKa = 4.05KK9 pKa = 9.82VFQSQARR16 pKa = 11.84RR17 pKa = 11.84FHH19 pKa = 6.42IVANEE24 pKa = 3.56KK25 pKa = 10.49SLPYY29 pKa = 10.49LNNILNFLTTKK40 pKa = 9.46PYY42 pKa = 11.07VYY44 pKa = 10.01ILCRR48 pKa = 11.84KK49 pKa = 9.54GYY51 pKa = 9.5NKK53 pKa = 10.14RR54 pKa = 11.84NCEE57 pKa = 3.7HH58 pKa = 5.95AHH60 pKa = 6.25IYY62 pKa = 10.76VIFKK66 pKa = 10.33KK67 pKa = 10.42AVRR70 pKa = 11.84LSNKK74 pKa = 7.36NTHH77 pKa = 5.8GCHH80 pKa = 5.69IAKK83 pKa = 10.09CRR85 pKa = 11.84GTTQQNIEE93 pKa = 4.59YY94 pKa = 9.57IKK96 pKa = 9.95GHH98 pKa = 6.53HH99 pKa = 6.29PMEE102 pKa = 4.42VCEE105 pKa = 4.36KK106 pKa = 11.38GEE108 pKa = 3.91IPEE111 pKa = 4.3NDD113 pKa = 3.27GDD115 pKa = 3.88IPPRR119 pKa = 11.84KK120 pKa = 8.56WDD122 pKa = 3.69GFVQQIKK129 pKa = 9.33QKK131 pKa = 10.69KK132 pKa = 7.94IDD134 pKa = 3.59KK135 pKa = 9.12FDD137 pKa = 3.63PMFARR142 pKa = 11.84YY143 pKa = 8.88EE144 pKa = 4.36GYY146 pKa = 10.61ANRR149 pKa = 11.84RR150 pKa = 11.84VAEE153 pKa = 3.95IMVEE157 pKa = 3.94EE158 pKa = 4.54LEE160 pKa = 5.51DD161 pKa = 3.95YY162 pKa = 11.19EE163 pKa = 7.06GDD165 pKa = 4.93LKK167 pKa = 11.35DD168 pKa = 3.89KK169 pKa = 11.04NCWIYY174 pKa = 11.25GEE176 pKa = 4.61PGTGKK181 pKa = 10.19SYY183 pKa = 10.56SARR186 pKa = 11.84YY187 pKa = 8.61CKK189 pKa = 9.98RR190 pKa = 11.84SKK192 pKa = 10.18IYY194 pKa = 9.41PKK196 pKa = 10.14PVSKK200 pKa = 9.54WWDD203 pKa = 3.5GYY205 pKa = 10.74KK206 pKa = 9.9GQKK209 pKa = 8.48VVVMDD214 pKa = 5.31DD215 pKa = 4.23LDD217 pKa = 4.1PDD219 pKa = 4.0RR220 pKa = 11.84AKK222 pKa = 10.46MLADD226 pKa = 4.83KK227 pKa = 10.84IKK229 pKa = 9.91IWTDD233 pKa = 3.0KK234 pKa = 10.55YY235 pKa = 10.86PFVAEE240 pKa = 4.47VKK242 pKa = 10.34SSSAVVDD249 pKa = 3.36PRR251 pKa = 11.84INFIVTSNYY260 pKa = 10.33SIEE263 pKa = 4.36DD264 pKa = 3.64CFEE267 pKa = 4.5SKK269 pKa = 10.57DD270 pKa = 3.4VEE272 pKa = 4.43AIKK275 pKa = 10.7RR276 pKa = 11.84RR277 pKa = 11.84FTVLHH282 pKa = 5.42FTGEE286 pKa = 4.39SIEE289 pKa = 4.25EE290 pKa = 4.06EE291 pKa = 4.2
Molecular weight: 34.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2L0HH22|A0A2L0HH22_9VIRU Replication-associated protein OS=Cattle blood-associated circovirus-like virus OX=2077298 PE=4 SV=1
MM1 pKa = 7.21SRR3 pKa = 11.84RR4 pKa = 11.84YY5 pKa = 8.56YY6 pKa = 10.44SRR8 pKa = 11.84RR9 pKa = 11.84TRR11 pKa = 11.84VIRR14 pKa = 11.84PKK16 pKa = 10.64KK17 pKa = 8.9KK18 pKa = 7.52WASNFRR24 pKa = 11.84QLTGLTSATVNVRR37 pKa = 11.84PKK39 pKa = 9.86SQAASTHH46 pKa = 7.08LIRR49 pKa = 11.84VWYY52 pKa = 8.28ILKK55 pKa = 10.6VKK57 pKa = 10.66GDD59 pKa = 3.42MRR61 pKa = 11.84LYY63 pKa = 8.92ITASSSIFSAQFASCVAYY81 pKa = 9.82IMYY84 pKa = 10.12LPEE87 pKa = 4.26GAAASIEE94 pKa = 4.45MIQSHH99 pKa = 6.56PEE101 pKa = 3.55WIMAWSPINVNMGTVSTSQPQDD123 pKa = 3.21QNVEE127 pKa = 4.23KK128 pKa = 10.74FSMSSRR134 pKa = 11.84LKK136 pKa = 10.69RR137 pKa = 11.84NLNSGDD143 pKa = 4.03KK144 pKa = 10.17IILVIDD150 pKa = 3.9LVFTPEE156 pKa = 3.77PTEE159 pKa = 4.06PSEE162 pKa = 4.17VPLNISYY169 pKa = 10.48QPAIGVQYY177 pKa = 7.36WTCANN182 pKa = 3.56
MM1 pKa = 7.21SRR3 pKa = 11.84RR4 pKa = 11.84YY5 pKa = 8.56YY6 pKa = 10.44SRR8 pKa = 11.84RR9 pKa = 11.84TRR11 pKa = 11.84VIRR14 pKa = 11.84PKK16 pKa = 10.64KK17 pKa = 8.9KK18 pKa = 7.52WASNFRR24 pKa = 11.84QLTGLTSATVNVRR37 pKa = 11.84PKK39 pKa = 9.86SQAASTHH46 pKa = 7.08LIRR49 pKa = 11.84VWYY52 pKa = 8.28ILKK55 pKa = 10.6VKK57 pKa = 10.66GDD59 pKa = 3.42MRR61 pKa = 11.84LYY63 pKa = 8.92ITASSSIFSAQFASCVAYY81 pKa = 9.82IMYY84 pKa = 10.12LPEE87 pKa = 4.26GAAASIEE94 pKa = 4.45MIQSHH99 pKa = 6.56PEE101 pKa = 3.55WIMAWSPINVNMGTVSTSQPQDD123 pKa = 3.21QNVEE127 pKa = 4.23KK128 pKa = 10.74FSMSSRR134 pKa = 11.84LKK136 pKa = 10.69RR137 pKa = 11.84NLNSGDD143 pKa = 4.03KK144 pKa = 10.17IILVIDD150 pKa = 3.9LVFTPEE156 pKa = 3.77PTEE159 pKa = 4.06PSEE162 pKa = 4.17VPLNISYY169 pKa = 10.48QPAIGVQYY177 pKa = 7.36WTCANN182 pKa = 3.56
Molecular weight: 20.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
473 |
182 |
291 |
236.5 |
27.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.92 ± 0.97 | 2.114 ± 0.555 |
4.651 ± 1.342 | 6.342 ± 1.366 |
3.594 ± 0.463 | 4.44 ± 0.625 |
2.114 ± 0.555 | 8.034 ± 0.114 |
8.879 ± 2.152 | 4.651 ± 0.762 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.748 ± 0.601 | 4.863 ± 0.045 |
5.074 ± 0.531 | 4.017 ± 0.508 |
5.92 ± 0.369 | 8.034 ± 2.218 |
4.44 ± 0.878 | 6.977 ± 0.091 |
2.114 ± 0.346 | 5.074 ± 0.371 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
