
Kwatta virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sunrhavirus; Kwatta sunrhavirus
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
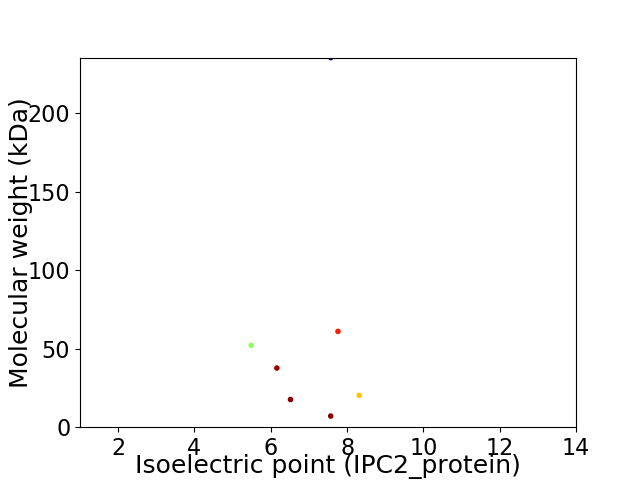
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D3R1B3|A0A0D3R1B3_9RHAB Phosphoprotein OS=Kwatta virus OX=1272945 PE=4 SV=1
MM1 pKa = 7.7PLSIVSTLSGKK12 pKa = 10.16KK13 pKa = 9.38VDD15 pKa = 4.01PLVVEE20 pKa = 4.62DD21 pKa = 5.47KK22 pKa = 11.18EE23 pKa = 4.13PLEE26 pKa = 4.2YY27 pKa = 9.76PSKK30 pKa = 10.82YY31 pKa = 8.8FTIHH35 pKa = 6.69SEE37 pKa = 3.46ITIFKK42 pKa = 8.83VQHH45 pKa = 5.09SLKK48 pKa = 10.69LNALAGGLIAQLNASVGSVEE68 pKa = 4.14GARR71 pKa = 11.84AFLTDD76 pKa = 3.6YY77 pKa = 11.03VSQFTQKK84 pKa = 10.69LSEE87 pKa = 4.29DD88 pKa = 3.25WTSFGVPLGRR98 pKa = 11.84ADD100 pKa = 4.03AEE102 pKa = 4.28VHH104 pKa = 5.7AFSYY108 pKa = 9.21VAWKK112 pKa = 9.97EE113 pKa = 4.12GPDD116 pKa = 3.27KK117 pKa = 10.8YY118 pKa = 8.31TTSGGNPVDD127 pKa = 4.0DD128 pKa = 5.43EE129 pKa = 5.16KK130 pKa = 11.74DD131 pKa = 3.79LLCLAVYY138 pKa = 10.38LCTVYY143 pKa = 10.79RR144 pKa = 11.84IGRR147 pKa = 11.84TSIAEE152 pKa = 3.71YY153 pKa = 8.78RR154 pKa = 11.84TRR156 pKa = 11.84LVAAGDD162 pKa = 3.61TFIRR166 pKa = 11.84GIKK169 pKa = 10.32ANFVGLDD176 pKa = 3.39NCLDD180 pKa = 3.67FCDD183 pKa = 5.58RR184 pKa = 11.84WVQDD188 pKa = 3.2QNYY191 pKa = 8.72CKK193 pKa = 10.08IVACIDD199 pKa = 3.48LFFKK203 pKa = 10.67KK204 pKa = 10.66FKK206 pKa = 10.23NHH208 pKa = 6.85DD209 pKa = 3.07IARR212 pKa = 11.84IRR214 pKa = 11.84FGTVAARR221 pKa = 11.84FRR223 pKa = 11.84EE224 pKa = 4.58CTALLSLAHH233 pKa = 5.76YY234 pKa = 10.04QDD236 pKa = 3.79LVGMKK241 pKa = 9.6IEE243 pKa = 4.37QALRR247 pKa = 11.84WIWIPRR253 pKa = 11.84VAEE256 pKa = 4.07QINNLMMPGQEE267 pKa = 4.15VLDD270 pKa = 4.08VTSLMPYY277 pKa = 10.11MMDD280 pKa = 3.51LGLSKK285 pKa = 10.67RR286 pKa = 11.84SPYY289 pKa = 10.29SAASNPEE296 pKa = 2.84WHH298 pKa = 6.94MYY300 pKa = 8.39VHH302 pKa = 7.38FIGALMGFKK311 pKa = 10.26RR312 pKa = 11.84SVDD315 pKa = 3.26ARR317 pKa = 11.84SLMVSGEE324 pKa = 4.2SNIMGNALVVAFVCRR339 pKa = 11.84SRR341 pKa = 11.84LTMEE345 pKa = 4.37RR346 pKa = 11.84MYY348 pKa = 9.68ITQEE352 pKa = 3.42EE353 pKa = 4.69HH354 pKa = 5.81EE355 pKa = 4.33AVEE358 pKa = 5.54KK359 pKa = 10.71KK360 pKa = 10.07RR361 pKa = 11.84QQNASAAAALPTKK374 pKa = 10.62SGTQEE379 pKa = 3.67KK380 pKa = 10.43AKK382 pKa = 10.43KK383 pKa = 9.86QEE385 pKa = 3.88EE386 pKa = 4.46TKK388 pKa = 11.08DD389 pKa = 3.61DD390 pKa = 4.31DD391 pKa = 4.26SDD393 pKa = 4.35AEE395 pKa = 4.23EE396 pKa = 4.07EE397 pKa = 4.67SEE399 pKa = 4.79FEE401 pKa = 4.13VDD403 pKa = 3.01ITGLPEE409 pKa = 4.12TQDD412 pKa = 2.94PDD414 pKa = 2.85MWFFFMKK421 pKa = 10.39GEE423 pKa = 4.17KK424 pKa = 9.94FRR426 pKa = 11.84IPDD429 pKa = 4.97AIYY432 pKa = 10.39DD433 pKa = 3.6WGTFRR438 pKa = 11.84AKK440 pKa = 10.55AIASTIRR447 pKa = 11.84KK448 pKa = 8.88GSVGEE453 pKa = 3.99RR454 pKa = 11.84LVLTFLGAQEE464 pKa = 3.93
MM1 pKa = 7.7PLSIVSTLSGKK12 pKa = 10.16KK13 pKa = 9.38VDD15 pKa = 4.01PLVVEE20 pKa = 4.62DD21 pKa = 5.47KK22 pKa = 11.18EE23 pKa = 4.13PLEE26 pKa = 4.2YY27 pKa = 9.76PSKK30 pKa = 10.82YY31 pKa = 8.8FTIHH35 pKa = 6.69SEE37 pKa = 3.46ITIFKK42 pKa = 8.83VQHH45 pKa = 5.09SLKK48 pKa = 10.69LNALAGGLIAQLNASVGSVEE68 pKa = 4.14GARR71 pKa = 11.84AFLTDD76 pKa = 3.6YY77 pKa = 11.03VSQFTQKK84 pKa = 10.69LSEE87 pKa = 4.29DD88 pKa = 3.25WTSFGVPLGRR98 pKa = 11.84ADD100 pKa = 4.03AEE102 pKa = 4.28VHH104 pKa = 5.7AFSYY108 pKa = 9.21VAWKK112 pKa = 9.97EE113 pKa = 4.12GPDD116 pKa = 3.27KK117 pKa = 10.8YY118 pKa = 8.31TTSGGNPVDD127 pKa = 4.0DD128 pKa = 5.43EE129 pKa = 5.16KK130 pKa = 11.74DD131 pKa = 3.79LLCLAVYY138 pKa = 10.38LCTVYY143 pKa = 10.79RR144 pKa = 11.84IGRR147 pKa = 11.84TSIAEE152 pKa = 3.71YY153 pKa = 8.78RR154 pKa = 11.84TRR156 pKa = 11.84LVAAGDD162 pKa = 3.61TFIRR166 pKa = 11.84GIKK169 pKa = 10.32ANFVGLDD176 pKa = 3.39NCLDD180 pKa = 3.67FCDD183 pKa = 5.58RR184 pKa = 11.84WVQDD188 pKa = 3.2QNYY191 pKa = 8.72CKK193 pKa = 10.08IVACIDD199 pKa = 3.48LFFKK203 pKa = 10.67KK204 pKa = 10.66FKK206 pKa = 10.23NHH208 pKa = 6.85DD209 pKa = 3.07IARR212 pKa = 11.84IRR214 pKa = 11.84FGTVAARR221 pKa = 11.84FRR223 pKa = 11.84EE224 pKa = 4.58CTALLSLAHH233 pKa = 5.76YY234 pKa = 10.04QDD236 pKa = 3.79LVGMKK241 pKa = 9.6IEE243 pKa = 4.37QALRR247 pKa = 11.84WIWIPRR253 pKa = 11.84VAEE256 pKa = 4.07QINNLMMPGQEE267 pKa = 4.15VLDD270 pKa = 4.08VTSLMPYY277 pKa = 10.11MMDD280 pKa = 3.51LGLSKK285 pKa = 10.67RR286 pKa = 11.84SPYY289 pKa = 10.29SAASNPEE296 pKa = 2.84WHH298 pKa = 6.94MYY300 pKa = 8.39VHH302 pKa = 7.38FIGALMGFKK311 pKa = 10.26RR312 pKa = 11.84SVDD315 pKa = 3.26ARR317 pKa = 11.84SLMVSGEE324 pKa = 4.2SNIMGNALVVAFVCRR339 pKa = 11.84SRR341 pKa = 11.84LTMEE345 pKa = 4.37RR346 pKa = 11.84MYY348 pKa = 9.68ITQEE352 pKa = 3.42EE353 pKa = 4.69HH354 pKa = 5.81EE355 pKa = 4.33AVEE358 pKa = 5.54KK359 pKa = 10.71KK360 pKa = 10.07RR361 pKa = 11.84QQNASAAAALPTKK374 pKa = 10.62SGTQEE379 pKa = 3.67KK380 pKa = 10.43AKK382 pKa = 10.43KK383 pKa = 9.86QEE385 pKa = 3.88EE386 pKa = 4.46TKK388 pKa = 11.08DD389 pKa = 3.61DD390 pKa = 4.31DD391 pKa = 4.26SDD393 pKa = 4.35AEE395 pKa = 4.23EE396 pKa = 4.07EE397 pKa = 4.67SEE399 pKa = 4.79FEE401 pKa = 4.13VDD403 pKa = 3.01ITGLPEE409 pKa = 4.12TQDD412 pKa = 2.94PDD414 pKa = 2.85MWFFFMKK421 pKa = 10.39GEE423 pKa = 4.17KK424 pKa = 9.94FRR426 pKa = 11.84IPDD429 pKa = 4.97AIYY432 pKa = 10.39DD433 pKa = 3.6WGTFRR438 pKa = 11.84AKK440 pKa = 10.55AIASTIRR447 pKa = 11.84KK448 pKa = 8.88GSVGEE453 pKa = 3.99RR454 pKa = 11.84LVLTFLGAQEE464 pKa = 3.93
Molecular weight: 52.19 kDa
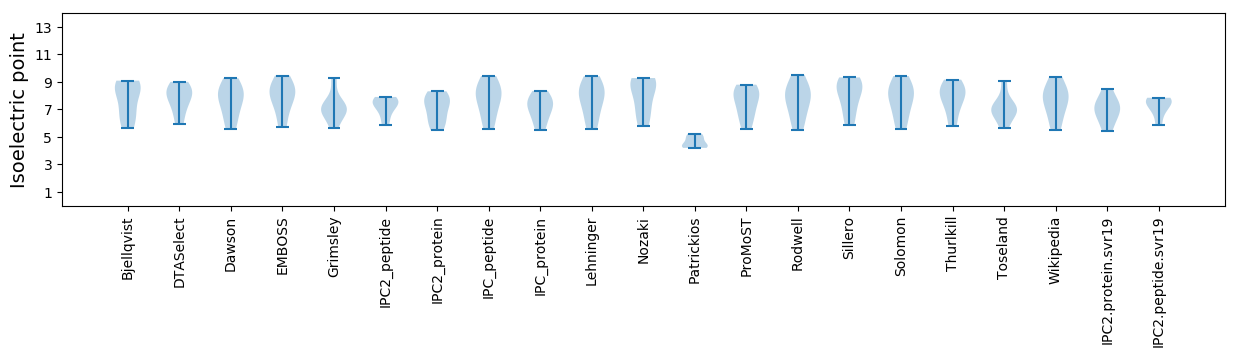
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D3R130|A0A0D3R130_9RHAB Nucleocapsid protein OS=Kwatta virus OX=1272945 PE=4 SV=1
MM1 pKa = 7.45ALFWTMFRR9 pKa = 11.84GKK11 pKa = 10.1RR12 pKa = 11.84DD13 pKa = 3.54DD14 pKa = 3.86YY15 pKa = 11.49FDD17 pKa = 4.45MGPAPQKK24 pKa = 9.67RR25 pKa = 11.84DD26 pKa = 3.41KK27 pKa = 11.67VNVKK31 pKa = 10.23CFVDD35 pKa = 4.19VSHH38 pKa = 7.83DD39 pKa = 3.24ISKK42 pKa = 8.84LTRR45 pKa = 11.84GITSDD50 pKa = 3.63EE51 pKa = 4.02LTNPIFPRR59 pKa = 11.84IDD61 pKa = 3.44CPLEE65 pKa = 3.69IRR67 pKa = 11.84SLVIYY72 pKa = 10.05LVGSIKK78 pKa = 9.72TYY80 pKa = 9.97RR81 pKa = 11.84RR82 pKa = 11.84KK83 pKa = 10.34KK84 pKa = 9.4EE85 pKa = 4.11DD86 pKa = 3.39KK87 pKa = 10.35TSLSLTWSILNLEE100 pKa = 4.47IPTDD104 pKa = 3.5RR105 pKa = 11.84PARR108 pKa = 11.84FDD110 pKa = 3.96EE111 pKa = 4.17TWKK114 pKa = 9.54WSKK117 pKa = 9.54TFSSYY122 pKa = 10.56HH123 pKa = 5.33GSEE126 pKa = 3.95PVFTNITVRR135 pKa = 11.84IEE137 pKa = 3.89PTSGKK142 pKa = 8.11GTSMDD147 pKa = 3.25VALLRR152 pKa = 11.84RR153 pKa = 11.84LEE155 pKa = 4.17SSLPNTQLIEE165 pKa = 4.24KK166 pKa = 10.01NEE168 pKa = 4.13DD169 pKa = 3.24GRR171 pKa = 11.84WQLVV175 pKa = 3.09
MM1 pKa = 7.45ALFWTMFRR9 pKa = 11.84GKK11 pKa = 10.1RR12 pKa = 11.84DD13 pKa = 3.54DD14 pKa = 3.86YY15 pKa = 11.49FDD17 pKa = 4.45MGPAPQKK24 pKa = 9.67RR25 pKa = 11.84DD26 pKa = 3.41KK27 pKa = 11.67VNVKK31 pKa = 10.23CFVDD35 pKa = 4.19VSHH38 pKa = 7.83DD39 pKa = 3.24ISKK42 pKa = 8.84LTRR45 pKa = 11.84GITSDD50 pKa = 3.63EE51 pKa = 4.02LTNPIFPRR59 pKa = 11.84IDD61 pKa = 3.44CPLEE65 pKa = 3.69IRR67 pKa = 11.84SLVIYY72 pKa = 10.05LVGSIKK78 pKa = 9.72TYY80 pKa = 9.97RR81 pKa = 11.84RR82 pKa = 11.84KK83 pKa = 10.34KK84 pKa = 9.4EE85 pKa = 4.11DD86 pKa = 3.39KK87 pKa = 10.35TSLSLTWSILNLEE100 pKa = 4.47IPTDD104 pKa = 3.5RR105 pKa = 11.84PARR108 pKa = 11.84FDD110 pKa = 3.96EE111 pKa = 4.17TWKK114 pKa = 9.54WSKK117 pKa = 9.54TFSSYY122 pKa = 10.56HH123 pKa = 5.33GSEE126 pKa = 3.95PVFTNITVRR135 pKa = 11.84IEE137 pKa = 3.89PTSGKK142 pKa = 8.11GTSMDD147 pKa = 3.25VALLRR152 pKa = 11.84RR153 pKa = 11.84LEE155 pKa = 4.17SSLPNTQLIEE165 pKa = 4.24KK166 pKa = 10.01NEE168 pKa = 4.13DD169 pKa = 3.24GRR171 pKa = 11.84WQLVV175 pKa = 3.09
Molecular weight: 20.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3803 |
63 |
2067 |
543.3 |
61.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.076 ± 0.885 | 1.972 ± 0.315 |
5.811 ± 0.336 | 5.969 ± 0.32 |
4.575 ± 0.451 | 6.179 ± 0.566 |
2.104 ± 0.244 | 6.968 ± 0.84 |
7.047 ± 0.364 | 10.465 ± 0.841 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.524 ± 0.201 | 4.391 ± 0.575 |
5.259 ± 1.366 | 3.55 ± 0.333 |
5.338 ± 0.537 | 7.915 ± 0.483 |
5.916 ± 0.286 | 5.653 ± 0.639 |
1.525 ± 0.196 | 2.761 ± 0.384 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
