
Pseudobacteroides cellulosolvens ATCC 35603 = DSM 2933
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Pseudobacteroides; Pseudobacteroides cellulosolvens
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
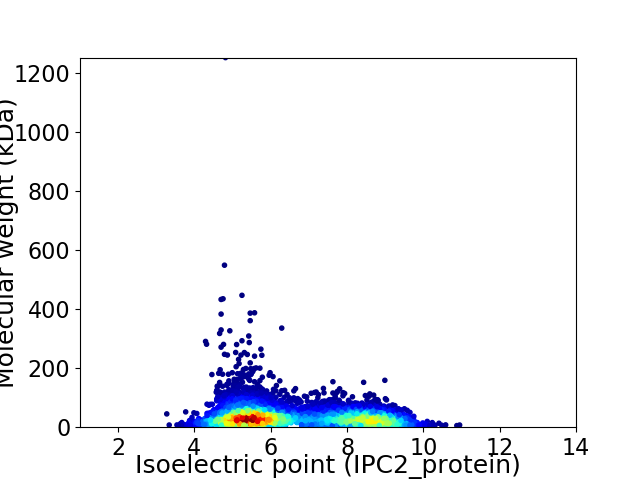
Virtual 2D-PAGE plot for 5771 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0L6JK86|A0A0L6JK86_9FIRM SH3 domain protein OS=Pseudobacteroides cellulosolvens ATCC 35603 = DSM 2933 OX=398512 GN=Bccel_1437 PE=4 SV=1
MM1 pKa = 7.64EE2 pKa = 5.36KK3 pKa = 10.28SCLGRR8 pKa = 11.84ALQTYY13 pKa = 9.67DD14 pKa = 3.86RR15 pKa = 11.84NNVSPKK21 pKa = 10.27SIQNKK26 pKa = 6.95TNSVEE31 pKa = 4.08EE32 pKa = 4.21VNMEE36 pKa = 4.27EE37 pKa = 4.09EE38 pKa = 4.19NKK40 pKa = 10.25LQEE43 pKa = 4.49EE44 pKa = 4.45IFPNEE49 pKa = 3.87NVVVEE54 pKa = 4.76LEE56 pKa = 4.02NSYY59 pKa = 10.68PEE61 pKa = 4.26PDD63 pKa = 2.94ISEE66 pKa = 4.24SMKK69 pKa = 11.05EE70 pKa = 3.77EE71 pKa = 3.98LQEE74 pKa = 4.09NTSEE78 pKa = 4.25STDD81 pKa = 3.69EE82 pKa = 4.26INLKK86 pKa = 9.09TFMEE90 pKa = 4.28NTMKK94 pKa = 10.61EE95 pKa = 4.11NEE97 pKa = 3.82IHH99 pKa = 5.94KK100 pKa = 9.46TNIYY104 pKa = 9.69EE105 pKa = 4.05NSKK108 pKa = 9.94KK109 pKa = 10.15IHH111 pKa = 6.01WNEE114 pKa = 3.66NMLKK118 pKa = 9.54RR119 pKa = 11.84CLGAYY124 pKa = 9.89YY125 pKa = 10.37SASQNTKK132 pKa = 8.22KK133 pKa = 8.62TRR135 pKa = 11.84KK136 pKa = 5.43TWKK139 pKa = 9.41RR140 pKa = 11.84KK141 pKa = 8.39EE142 pKa = 4.23SNICDD147 pKa = 3.48EE148 pKa = 4.46NVVSKK153 pKa = 11.23NEE155 pKa = 3.85EE156 pKa = 4.48LISSEE161 pKa = 3.78SSYY164 pKa = 11.28DD165 pKa = 3.32IVPQEE170 pKa = 3.97DD171 pKa = 3.65HH172 pKa = 6.8MLDD175 pKa = 3.44SQIYY179 pKa = 8.49TGEE182 pKa = 4.01TTEE185 pKa = 5.07KK186 pKa = 10.36VNEE189 pKa = 4.03EE190 pKa = 3.75NEE192 pKa = 4.51YY193 pKa = 10.44YY194 pKa = 10.75YY195 pKa = 10.37STGSEE200 pKa = 4.39SNNDD204 pKa = 3.13EE205 pKa = 3.9SDD207 pKa = 3.66EE208 pKa = 4.24NEE210 pKa = 3.91KK211 pKa = 11.06SEE213 pKa = 5.32GEE215 pKa = 4.02FTYY218 pKa = 10.97SSDD221 pKa = 3.56NEE223 pKa = 4.48SYY225 pKa = 10.88EE226 pKa = 3.78QDD228 pKa = 3.32SLGKK232 pKa = 9.85FEE234 pKa = 5.27KK235 pKa = 10.29ISWSIISDD243 pKa = 3.35IDD245 pKa = 3.49EE246 pKa = 4.12EE247 pKa = 5.84DD248 pKa = 3.7YY249 pKa = 11.41DD250 pKa = 5.27GEE252 pKa = 4.22MDD254 pKa = 6.15SIDD257 pKa = 4.9EE258 pKa = 5.52DD259 pKa = 3.47IQADD263 pKa = 3.82NEE265 pKa = 4.56SSNEE269 pKa = 3.75TDD271 pKa = 3.09SYY273 pKa = 11.23EE274 pKa = 4.07GRR276 pKa = 11.84SDD278 pKa = 3.55VADD281 pKa = 3.8SYY283 pKa = 12.15GGGSDD288 pKa = 5.21DD289 pKa = 4.23ADD291 pKa = 3.81SYY293 pKa = 11.7EE294 pKa = 4.68DD295 pKa = 4.41GYY297 pKa = 11.58EE298 pKa = 3.78NDD300 pKa = 4.03YY301 pKa = 11.07EE302 pKa = 4.15DD303 pKa = 5.8SYY305 pKa = 11.38VQEE308 pKa = 5.39DD309 pKa = 3.98LLKK312 pKa = 10.88DD313 pKa = 3.36KK314 pKa = 10.98YY315 pKa = 10.99HH316 pKa = 6.58NGNNGTDD323 pKa = 3.33QYY325 pKa = 11.33FSEE328 pKa = 4.76AVDD331 pKa = 4.69DD332 pKa = 4.52NFEE335 pKa = 4.03SDD337 pKa = 3.8YY338 pKa = 11.46QEE340 pKa = 4.71NEE342 pKa = 3.58VLAYY346 pKa = 10.36SDD348 pKa = 3.79EE349 pKa = 4.49NEE351 pKa = 4.31VYY353 pKa = 10.44DD354 pKa = 4.68FEE356 pKa = 5.8DD357 pKa = 3.37MRR359 pKa = 11.84PVEE362 pKa = 4.16TLDD365 pKa = 5.26LEE367 pKa = 4.49IANNTIDD374 pKa = 3.57NTNWDD379 pKa = 4.32NGWDD383 pKa = 3.66EE384 pKa = 5.25CDD386 pKa = 3.65VLNNEE391 pKa = 4.27YY392 pKa = 11.14NDD394 pKa = 3.61IEE396 pKa = 4.72TVGSFIDD403 pKa = 3.58YY404 pKa = 10.8RR405 pKa = 11.84NISEE409 pKa = 5.86DD410 pKa = 3.46YY411 pKa = 10.85DD412 pKa = 5.74DD413 pKa = 6.15YY414 pKa = 12.11DD415 pKa = 5.02FDD417 pKa = 4.38CQTQIEE423 pKa = 4.7TPEE426 pKa = 3.92YY427 pKa = 9.92MSDD430 pKa = 3.54YY431 pKa = 11.16DD432 pKa = 3.8QISYY436 pKa = 9.12GQSVEE441 pKa = 4.04CRR443 pKa = 11.84IITT446 pKa = 3.66
MM1 pKa = 7.64EE2 pKa = 5.36KK3 pKa = 10.28SCLGRR8 pKa = 11.84ALQTYY13 pKa = 9.67DD14 pKa = 3.86RR15 pKa = 11.84NNVSPKK21 pKa = 10.27SIQNKK26 pKa = 6.95TNSVEE31 pKa = 4.08EE32 pKa = 4.21VNMEE36 pKa = 4.27EE37 pKa = 4.09EE38 pKa = 4.19NKK40 pKa = 10.25LQEE43 pKa = 4.49EE44 pKa = 4.45IFPNEE49 pKa = 3.87NVVVEE54 pKa = 4.76LEE56 pKa = 4.02NSYY59 pKa = 10.68PEE61 pKa = 4.26PDD63 pKa = 2.94ISEE66 pKa = 4.24SMKK69 pKa = 11.05EE70 pKa = 3.77EE71 pKa = 3.98LQEE74 pKa = 4.09NTSEE78 pKa = 4.25STDD81 pKa = 3.69EE82 pKa = 4.26INLKK86 pKa = 9.09TFMEE90 pKa = 4.28NTMKK94 pKa = 10.61EE95 pKa = 4.11NEE97 pKa = 3.82IHH99 pKa = 5.94KK100 pKa = 9.46TNIYY104 pKa = 9.69EE105 pKa = 4.05NSKK108 pKa = 9.94KK109 pKa = 10.15IHH111 pKa = 6.01WNEE114 pKa = 3.66NMLKK118 pKa = 9.54RR119 pKa = 11.84CLGAYY124 pKa = 9.89YY125 pKa = 10.37SASQNTKK132 pKa = 8.22KK133 pKa = 8.62TRR135 pKa = 11.84KK136 pKa = 5.43TWKK139 pKa = 9.41RR140 pKa = 11.84KK141 pKa = 8.39EE142 pKa = 4.23SNICDD147 pKa = 3.48EE148 pKa = 4.46NVVSKK153 pKa = 11.23NEE155 pKa = 3.85EE156 pKa = 4.48LISSEE161 pKa = 3.78SSYY164 pKa = 11.28DD165 pKa = 3.32IVPQEE170 pKa = 3.97DD171 pKa = 3.65HH172 pKa = 6.8MLDD175 pKa = 3.44SQIYY179 pKa = 8.49TGEE182 pKa = 4.01TTEE185 pKa = 5.07KK186 pKa = 10.36VNEE189 pKa = 4.03EE190 pKa = 3.75NEE192 pKa = 4.51YY193 pKa = 10.44YY194 pKa = 10.75YY195 pKa = 10.37STGSEE200 pKa = 4.39SNNDD204 pKa = 3.13EE205 pKa = 3.9SDD207 pKa = 3.66EE208 pKa = 4.24NEE210 pKa = 3.91KK211 pKa = 11.06SEE213 pKa = 5.32GEE215 pKa = 4.02FTYY218 pKa = 10.97SSDD221 pKa = 3.56NEE223 pKa = 4.48SYY225 pKa = 10.88EE226 pKa = 3.78QDD228 pKa = 3.32SLGKK232 pKa = 9.85FEE234 pKa = 5.27KK235 pKa = 10.29ISWSIISDD243 pKa = 3.35IDD245 pKa = 3.49EE246 pKa = 4.12EE247 pKa = 5.84DD248 pKa = 3.7YY249 pKa = 11.41DD250 pKa = 5.27GEE252 pKa = 4.22MDD254 pKa = 6.15SIDD257 pKa = 4.9EE258 pKa = 5.52DD259 pKa = 3.47IQADD263 pKa = 3.82NEE265 pKa = 4.56SSNEE269 pKa = 3.75TDD271 pKa = 3.09SYY273 pKa = 11.23EE274 pKa = 4.07GRR276 pKa = 11.84SDD278 pKa = 3.55VADD281 pKa = 3.8SYY283 pKa = 12.15GGGSDD288 pKa = 5.21DD289 pKa = 4.23ADD291 pKa = 3.81SYY293 pKa = 11.7EE294 pKa = 4.68DD295 pKa = 4.41GYY297 pKa = 11.58EE298 pKa = 3.78NDD300 pKa = 4.03YY301 pKa = 11.07EE302 pKa = 4.15DD303 pKa = 5.8SYY305 pKa = 11.38VQEE308 pKa = 5.39DD309 pKa = 3.98LLKK312 pKa = 10.88DD313 pKa = 3.36KK314 pKa = 10.98YY315 pKa = 10.99HH316 pKa = 6.58NGNNGTDD323 pKa = 3.33QYY325 pKa = 11.33FSEE328 pKa = 4.76AVDD331 pKa = 4.69DD332 pKa = 4.52NFEE335 pKa = 4.03SDD337 pKa = 3.8YY338 pKa = 11.46QEE340 pKa = 4.71NEE342 pKa = 3.58VLAYY346 pKa = 10.36SDD348 pKa = 3.79EE349 pKa = 4.49NEE351 pKa = 4.31VYY353 pKa = 10.44DD354 pKa = 4.68FEE356 pKa = 5.8DD357 pKa = 3.37MRR359 pKa = 11.84PVEE362 pKa = 4.16TLDD365 pKa = 5.26LEE367 pKa = 4.49IANNTIDD374 pKa = 3.57NTNWDD379 pKa = 4.32NGWDD383 pKa = 3.66EE384 pKa = 5.25CDD386 pKa = 3.65VLNNEE391 pKa = 4.27YY392 pKa = 11.14NDD394 pKa = 3.61IEE396 pKa = 4.72TVGSFIDD403 pKa = 3.58YY404 pKa = 10.8RR405 pKa = 11.84NISEE409 pKa = 5.86DD410 pKa = 3.46YY411 pKa = 10.85DD412 pKa = 5.74DD413 pKa = 6.15YY414 pKa = 12.11DD415 pKa = 5.02FDD417 pKa = 4.38CQTQIEE423 pKa = 4.7TPEE426 pKa = 3.92YY427 pKa = 9.92MSDD430 pKa = 3.54YY431 pKa = 11.16DD432 pKa = 3.8QISYY436 pKa = 9.12GQSVEE441 pKa = 4.04CRR443 pKa = 11.84IITT446 pKa = 3.66
Molecular weight: 51.9 kDa
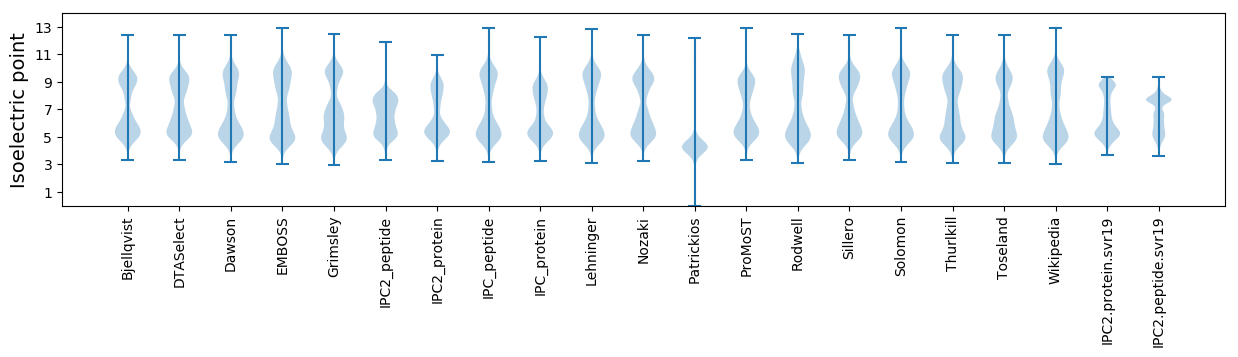
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0L6JX97|A0A0L6JX97_9FIRM Sulfate-transporting ATPase OS=Pseudobacteroides cellulosolvens ATCC 35603 = DSM 2933 OX=398512 GN=Bccel_5349 PE=4 SV=1
MM1 pKa = 7.69RR2 pKa = 11.84VLIAGSRR9 pKa = 11.84FYY11 pKa = 11.09RR12 pKa = 11.84DD13 pKa = 3.31YY14 pKa = 11.49QKK16 pKa = 10.8ILAVVRR22 pKa = 11.84SLDD25 pKa = 3.25IDD27 pKa = 3.8LVIAGGCRR35 pKa = 11.84VLMLWLFVLLVSAASNILSIRR56 pKa = 11.84QIGRR60 pKa = 11.84GLAKK64 pKa = 10.7VPDD67 pKa = 4.3LSGMPP72 pKa = 3.72
MM1 pKa = 7.69RR2 pKa = 11.84VLIAGSRR9 pKa = 11.84FYY11 pKa = 11.09RR12 pKa = 11.84DD13 pKa = 3.31YY14 pKa = 11.49QKK16 pKa = 10.8ILAVVRR22 pKa = 11.84SLDD25 pKa = 3.25IDD27 pKa = 3.8LVIAGGCRR35 pKa = 11.84VLMLWLFVLLVSAASNILSIRR56 pKa = 11.84QIGRR60 pKa = 11.84GLAKK64 pKa = 10.7VPDD67 pKa = 4.3LSGMPP72 pKa = 3.72
Molecular weight: 7.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1943841 |
30 |
11562 |
336.8 |
37.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.962 ± 0.033 | 1.19 ± 0.014 |
5.861 ± 0.028 | 6.505 ± 0.039 |
4.366 ± 0.025 | 6.567 ± 0.033 |
1.442 ± 0.014 | 8.963 ± 0.039 |
8.161 ± 0.032 | 8.6 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.569 ± 0.019 | 6.034 ± 0.034 |
3.326 ± 0.025 | 2.668 ± 0.017 |
3.588 ± 0.026 | 7.02 ± 0.037 |
5.442 ± 0.053 | 6.52 ± 0.029 |
0.962 ± 0.013 | 4.254 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
