
Polaribacter atrinae
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Polaribacter
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
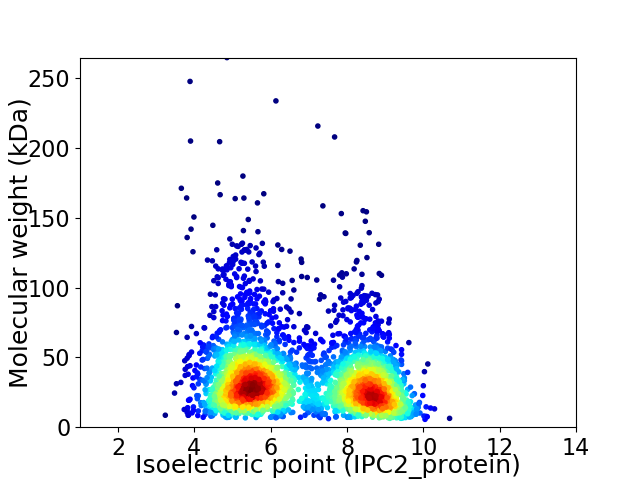
Virtual 2D-PAGE plot for 3258 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A176TEQ9|A0A176TEQ9_9FLAO Methionine--tRNA ligase OS=Polaribacter atrinae OX=1333662 GN=metG PE=3 SV=1
MM1 pKa = 7.71LFFCFITFQNINAQLSKK18 pKa = 10.91KK19 pKa = 10.07HH20 pKa = 6.51FIPPLTSTDD29 pKa = 3.33GFTDD33 pKa = 3.12QYY35 pKa = 11.33IYY37 pKa = 10.74ISTPNNNVSYY47 pKa = 10.72KK48 pKa = 8.61ITPVGNLDD56 pKa = 3.17QTAYY60 pKa = 10.84SGIVSNNLPIEE71 pKa = 4.1QPVLNLSGNTDD82 pKa = 4.07FDD84 pKa = 4.44NDD86 pKa = 3.96TQLHH90 pKa = 5.86IPEE93 pKa = 4.59SSIIGNKK100 pKa = 6.03FTNRR104 pKa = 11.84GFIIEE109 pKa = 4.62ANDD112 pKa = 4.46LIYY115 pKa = 10.27VTIRR119 pKa = 11.84VRR121 pKa = 11.84SSQANNGNKK130 pKa = 8.8NQAGALVSKK139 pKa = 10.59GNSALGTEE147 pKa = 4.37FRR149 pKa = 11.84IGGFIRR155 pKa = 11.84DD156 pKa = 3.58GSINGDD162 pKa = 3.46VTFASIMATEE172 pKa = 5.18DD173 pKa = 3.29NTEE176 pKa = 3.97VTITNATSSSTIININLNEE195 pKa = 4.25GEE197 pKa = 4.53SYY199 pKa = 10.43IIHH202 pKa = 7.07ANGANSSSIIGNLITSDD219 pKa = 3.32KK220 pKa = 10.25PIVVNSGSAYY230 pKa = 10.37GSFGNNSGNADD241 pKa = 3.59YY242 pKa = 11.23GFDD245 pKa = 3.84QIVGADD251 pKa = 3.93KK252 pKa = 10.78IGSEE256 pKa = 4.45YY257 pKa = 10.6ILVKK261 pKa = 10.86GSGNTIEE268 pKa = 4.38KK269 pKa = 10.46SLIIAHH275 pKa = 6.68EE276 pKa = 4.48DD277 pKa = 3.14FTEE280 pKa = 4.41FFINDD285 pKa = 3.49TSVGTFNAGEE295 pKa = 4.18YY296 pKa = 10.66YY297 pKa = 9.24ITTNSMYY304 pKa = 7.63TTNGNMYY311 pKa = 10.19INTNKK316 pKa = 9.96PVFVYY321 pKa = 10.32QGVGGLGNNGIPSEE335 pKa = 4.16ANQGLFFVPPLSCEE349 pKa = 3.84NRR351 pKa = 11.84GKK353 pKa = 10.63VDD355 pKa = 4.67NIPHH359 pKa = 6.73IEE361 pKa = 4.25RR362 pKa = 11.84IGNEE366 pKa = 4.18DD367 pKa = 3.26FTGGISIVTNEE378 pKa = 4.17DD379 pKa = 3.44SVVKK383 pKa = 10.67INGSLITTFSGISGPFLITTKK404 pKa = 10.93DD405 pKa = 2.99SGKK408 pKa = 9.67YY409 pKa = 8.48DD410 pKa = 3.58TYY412 pKa = 11.42KK413 pKa = 9.78VTGLEE418 pKa = 4.18GNISIEE424 pKa = 4.2SSGEE428 pKa = 3.83LYY430 pKa = 10.5CAYY433 pKa = 10.46FNQNGLATSGAFYY446 pKa = 10.75SGFPSSPEE454 pKa = 3.33IKK456 pKa = 10.37FNVAISTLGNCLGNDD471 pKa = 4.52LEE473 pKa = 4.44LQAANTEE480 pKa = 4.39LFDD483 pKa = 3.9SFKK486 pKa = 10.57WYY488 pKa = 10.7FNDD491 pKa = 3.59GSGYY495 pKa = 9.83IDD497 pKa = 3.66TNEE500 pKa = 3.95TSATLIPSAPGSYY513 pKa = 10.4QLIGKK518 pKa = 8.73INCTNTTFKK527 pKa = 10.2STEE530 pKa = 4.0IPVSICPDD538 pKa = 3.88DD539 pKa = 3.97YY540 pKa = 11.91DD541 pKa = 5.05KK542 pKa = 11.78DD543 pKa = 4.35GIIDD547 pKa = 4.51NIDD550 pKa = 2.88VDD552 pKa = 3.81IDD554 pKa = 3.54NDD556 pKa = 4.61GILNIDD562 pKa = 3.73EE563 pKa = 4.43SLGDD567 pKa = 4.13KK568 pKa = 10.54IIDD571 pKa = 3.85LSDD574 pKa = 3.29INNPTVEE581 pKa = 4.86DD582 pKa = 4.25DD583 pKa = 3.55VTTIITNTFTGDD595 pKa = 2.82ATGNFTSVANNVTTSNATYY614 pKa = 10.92SLDD617 pKa = 3.64FNDD620 pKa = 4.86NINFKK625 pKa = 9.2LTQNRR630 pKa = 11.84TTDD633 pKa = 3.22HH634 pKa = 6.54TISNNEE640 pKa = 3.51YY641 pKa = 10.53FIIKK645 pKa = 10.46LSQTEE650 pKa = 4.13KK651 pKa = 10.94NITLIDD657 pKa = 4.36PDD659 pKa = 3.96NQILIDD665 pKa = 4.18SNFDD669 pKa = 5.1DD670 pKa = 4.23EE671 pKa = 5.88FEE673 pKa = 5.6DD674 pKa = 4.7NFTQFSASVIKK685 pKa = 10.49FKK687 pKa = 11.11FKK689 pKa = 11.04ADD691 pKa = 3.78LVGSTSTFQFVANQINNISFEE712 pKa = 4.28HH713 pKa = 6.49YY714 pKa = 11.01NDD716 pKa = 3.24ISSNASTFNGNIGLTCFSLDD736 pKa = 3.26SDD738 pKa = 4.01GDD740 pKa = 4.14GIEE743 pKa = 5.74DD744 pKa = 4.25MFDD747 pKa = 4.28LDD749 pKa = 4.25TDD751 pKa = 3.61NDD753 pKa = 5.31GIPDD757 pKa = 4.28LYY759 pKa = 10.87DD760 pKa = 3.89AYY762 pKa = 10.64GQEE765 pKa = 3.76ISLAKK770 pKa = 9.87TDD772 pKa = 4.09ANLDD776 pKa = 3.52GLDD779 pKa = 4.41DD780 pKa = 3.94VFEE783 pKa = 5.33SITTNLDD790 pKa = 3.03TDD792 pKa = 3.88GDD794 pKa = 4.35GVNNYY799 pKa = 9.9LDD801 pKa = 4.0YY802 pKa = 11.49DD803 pKa = 3.92SDD805 pKa = 3.92NDD807 pKa = 4.57GIYY810 pKa = 10.82DD811 pKa = 3.66STEE814 pKa = 3.73ASHH817 pKa = 7.07GLDD820 pKa = 2.95TDD822 pKa = 4.07FNGTIDD828 pKa = 3.64TFIDD832 pKa = 3.22INKK835 pKa = 10.02NGLEE839 pKa = 4.24DD840 pKa = 3.7SLEE843 pKa = 4.22NDD845 pKa = 3.56TTVQTISLKK854 pKa = 9.4YY855 pKa = 9.17TIADD859 pKa = 3.57TDD861 pKa = 3.37ADD863 pKa = 4.26AIFNFIEE870 pKa = 4.69LDD872 pKa = 3.63SDD874 pKa = 4.25ADD876 pKa = 3.67GCNDD880 pKa = 3.29VIEE883 pKa = 5.78AGFTDD888 pKa = 3.86NNNDD892 pKa = 3.73GILFANSFSVDD903 pKa = 3.15EE904 pKa = 4.39NGKK907 pKa = 9.76VKK909 pKa = 10.76NPNDD913 pKa = 4.15GYY915 pKa = 7.67TTPNSNYY922 pKa = 6.83TTSAPIVVTTFTDD935 pKa = 3.44VTFCEE940 pKa = 4.22NLTDD944 pKa = 5.71IITIDD949 pKa = 3.56SNADD953 pKa = 3.06SFQWEE958 pKa = 4.49FSTDD962 pKa = 3.07GTNWLIINEE971 pKa = 4.62DD972 pKa = 3.93DD973 pKa = 4.24TTDD976 pKa = 3.89LYY978 pKa = 11.25QGSTSASLQITNTPFTYY995 pKa = 10.23DD996 pKa = 2.83SYY998 pKa = 11.32QYY1000 pKa = 10.66RR1001 pKa = 11.84VALNRR1006 pKa = 11.84TGNSCGFTSDD1016 pKa = 5.5AITLTVNPKK1025 pKa = 9.91PVIEE1029 pKa = 4.54NNPAALFQCDD1039 pKa = 4.04TDD1041 pKa = 4.67SDD1043 pKa = 4.14LQTTFNLTEE1052 pKa = 4.17SEE1054 pKa = 3.84ISITNEE1060 pKa = 3.12TDD1062 pKa = 3.32VNFEE1066 pKa = 4.23YY1067 pKa = 11.09YY1068 pKa = 9.36ATEE1071 pKa = 3.93LDD1073 pKa = 4.88AINGTPQVADD1083 pKa = 3.37KK1084 pKa = 10.72TSYY1087 pKa = 10.81SVNTNGEE1094 pKa = 4.05AWVKK1098 pKa = 9.78TISNTTACYY1107 pKa = 10.27SISKK1111 pKa = 10.27INLTVSYY1118 pKa = 10.76APNEE1122 pKa = 4.34TFNDD1126 pKa = 3.41TLYY1129 pKa = 10.97QCDD1132 pKa = 4.82DD1133 pKa = 4.27LLDD1136 pKa = 4.59LDD1138 pKa = 5.57GNNTTNNDD1146 pKa = 3.72DD1147 pKa = 3.69KK1148 pKa = 11.81DD1149 pKa = 4.54GITSFDD1155 pKa = 3.94LSNISDD1161 pKa = 4.48KK1162 pKa = 11.02ISTDD1166 pKa = 2.96SDD1168 pKa = 3.28IKK1170 pKa = 10.82IEE1172 pKa = 4.72FYY1174 pKa = 10.16EE1175 pKa = 4.22TDD1177 pKa = 3.48DD1178 pKa = 4.74DD1179 pKa = 4.03RR1180 pKa = 11.84TRR1182 pKa = 11.84SLNEE1186 pKa = 3.26ITEE1189 pKa = 4.42TQDD1192 pKa = 3.08LASYY1196 pKa = 10.37RR1197 pKa = 11.84NKK1199 pKa = 10.51NYY1201 pKa = 10.09PFTTGNPVTIFYY1213 pKa = 10.91KK1214 pKa = 10.8LISITNNNCQGIGEE1228 pKa = 4.97FYY1230 pKa = 11.2LEE1232 pKa = 4.24VNKK1235 pKa = 10.29IPEE1238 pKa = 4.06FTVDD1242 pKa = 4.24GEE1244 pKa = 4.44APSDD1248 pKa = 4.22PIIICAEE1255 pKa = 4.21NIPYY1259 pKa = 8.67TLNVYY1264 pKa = 8.93TSNSSYY1270 pKa = 11.06NYY1272 pKa = 9.35QWTNEE1277 pKa = 4.03KK1278 pKa = 10.56GDD1280 pKa = 3.97NLGTTQNINISDD1292 pKa = 3.67AGVYY1296 pKa = 8.72TVTSSTNSCSRR1307 pKa = 11.84SRR1309 pKa = 11.84TITVYY1314 pKa = 10.79KK1315 pKa = 10.5SDD1317 pKa = 3.87FEE1319 pKa = 4.22NLEE1322 pKa = 4.03EE1323 pKa = 4.45SFVTITDD1330 pKa = 3.69DD1331 pKa = 4.61LSVINSNLNIRR1342 pKa = 11.84IDD1344 pKa = 3.45IPTNPLINEE1353 pKa = 4.3VFQYY1357 pKa = 11.39ALEE1360 pKa = 5.29DD1361 pKa = 3.81EE1362 pKa = 5.24DD1363 pKa = 5.17GMTIRR1368 pKa = 11.84SFQDD1372 pKa = 3.03SNVFEE1377 pKa = 5.49DD1378 pKa = 3.98IEE1380 pKa = 4.41GGIYY1384 pKa = 9.98KK1385 pKa = 10.17IIVEE1389 pKa = 4.48SKK1391 pKa = 9.03NGCGTSEE1398 pKa = 4.47LLVSVIQFPKK1408 pKa = 10.42FFTPNGDD1415 pKa = 3.34RR1416 pKa = 11.84KK1417 pKa = 10.33NDD1419 pKa = 2.79SWGIKK1424 pKa = 9.47GVSNTLYY1431 pKa = 10.49QSNSSINIFNRR1442 pKa = 11.84YY1443 pKa = 7.94GKK1445 pKa = 10.34LVAQTTIDD1453 pKa = 3.51GDD1455 pKa = 3.88GWDD1458 pKa = 3.44GTYY1461 pKa = 10.52NGKK1464 pKa = 9.2LLPSDD1469 pKa = 4.78DD1470 pKa = 3.67YY1471 pKa = 10.8WYY1473 pKa = 10.48SIQLIPVNTDD1483 pKa = 2.8QKK1485 pKa = 10.67PILKK1489 pKa = 9.71KK1490 pKa = 10.6GHH1492 pKa = 6.37FSLIRR1497 pKa = 11.84KK1498 pKa = 8.22
MM1 pKa = 7.71LFFCFITFQNINAQLSKK18 pKa = 10.91KK19 pKa = 10.07HH20 pKa = 6.51FIPPLTSTDD29 pKa = 3.33GFTDD33 pKa = 3.12QYY35 pKa = 11.33IYY37 pKa = 10.74ISTPNNNVSYY47 pKa = 10.72KK48 pKa = 8.61ITPVGNLDD56 pKa = 3.17QTAYY60 pKa = 10.84SGIVSNNLPIEE71 pKa = 4.1QPVLNLSGNTDD82 pKa = 4.07FDD84 pKa = 4.44NDD86 pKa = 3.96TQLHH90 pKa = 5.86IPEE93 pKa = 4.59SSIIGNKK100 pKa = 6.03FTNRR104 pKa = 11.84GFIIEE109 pKa = 4.62ANDD112 pKa = 4.46LIYY115 pKa = 10.27VTIRR119 pKa = 11.84VRR121 pKa = 11.84SSQANNGNKK130 pKa = 8.8NQAGALVSKK139 pKa = 10.59GNSALGTEE147 pKa = 4.37FRR149 pKa = 11.84IGGFIRR155 pKa = 11.84DD156 pKa = 3.58GSINGDD162 pKa = 3.46VTFASIMATEE172 pKa = 5.18DD173 pKa = 3.29NTEE176 pKa = 3.97VTITNATSSSTIININLNEE195 pKa = 4.25GEE197 pKa = 4.53SYY199 pKa = 10.43IIHH202 pKa = 7.07ANGANSSSIIGNLITSDD219 pKa = 3.32KK220 pKa = 10.25PIVVNSGSAYY230 pKa = 10.37GSFGNNSGNADD241 pKa = 3.59YY242 pKa = 11.23GFDD245 pKa = 3.84QIVGADD251 pKa = 3.93KK252 pKa = 10.78IGSEE256 pKa = 4.45YY257 pKa = 10.6ILVKK261 pKa = 10.86GSGNTIEE268 pKa = 4.38KK269 pKa = 10.46SLIIAHH275 pKa = 6.68EE276 pKa = 4.48DD277 pKa = 3.14FTEE280 pKa = 4.41FFINDD285 pKa = 3.49TSVGTFNAGEE295 pKa = 4.18YY296 pKa = 10.66YY297 pKa = 9.24ITTNSMYY304 pKa = 7.63TTNGNMYY311 pKa = 10.19INTNKK316 pKa = 9.96PVFVYY321 pKa = 10.32QGVGGLGNNGIPSEE335 pKa = 4.16ANQGLFFVPPLSCEE349 pKa = 3.84NRR351 pKa = 11.84GKK353 pKa = 10.63VDD355 pKa = 4.67NIPHH359 pKa = 6.73IEE361 pKa = 4.25RR362 pKa = 11.84IGNEE366 pKa = 4.18DD367 pKa = 3.26FTGGISIVTNEE378 pKa = 4.17DD379 pKa = 3.44SVVKK383 pKa = 10.67INGSLITTFSGISGPFLITTKK404 pKa = 10.93DD405 pKa = 2.99SGKK408 pKa = 9.67YY409 pKa = 8.48DD410 pKa = 3.58TYY412 pKa = 11.42KK413 pKa = 9.78VTGLEE418 pKa = 4.18GNISIEE424 pKa = 4.2SSGEE428 pKa = 3.83LYY430 pKa = 10.5CAYY433 pKa = 10.46FNQNGLATSGAFYY446 pKa = 10.75SGFPSSPEE454 pKa = 3.33IKK456 pKa = 10.37FNVAISTLGNCLGNDD471 pKa = 4.52LEE473 pKa = 4.44LQAANTEE480 pKa = 4.39LFDD483 pKa = 3.9SFKK486 pKa = 10.57WYY488 pKa = 10.7FNDD491 pKa = 3.59GSGYY495 pKa = 9.83IDD497 pKa = 3.66TNEE500 pKa = 3.95TSATLIPSAPGSYY513 pKa = 10.4QLIGKK518 pKa = 8.73INCTNTTFKK527 pKa = 10.2STEE530 pKa = 4.0IPVSICPDD538 pKa = 3.88DD539 pKa = 3.97YY540 pKa = 11.91DD541 pKa = 5.05KK542 pKa = 11.78DD543 pKa = 4.35GIIDD547 pKa = 4.51NIDD550 pKa = 2.88VDD552 pKa = 3.81IDD554 pKa = 3.54NDD556 pKa = 4.61GILNIDD562 pKa = 3.73EE563 pKa = 4.43SLGDD567 pKa = 4.13KK568 pKa = 10.54IIDD571 pKa = 3.85LSDD574 pKa = 3.29INNPTVEE581 pKa = 4.86DD582 pKa = 4.25DD583 pKa = 3.55VTTIITNTFTGDD595 pKa = 2.82ATGNFTSVANNVTTSNATYY614 pKa = 10.92SLDD617 pKa = 3.64FNDD620 pKa = 4.86NINFKK625 pKa = 9.2LTQNRR630 pKa = 11.84TTDD633 pKa = 3.22HH634 pKa = 6.54TISNNEE640 pKa = 3.51YY641 pKa = 10.53FIIKK645 pKa = 10.46LSQTEE650 pKa = 4.13KK651 pKa = 10.94NITLIDD657 pKa = 4.36PDD659 pKa = 3.96NQILIDD665 pKa = 4.18SNFDD669 pKa = 5.1DD670 pKa = 4.23EE671 pKa = 5.88FEE673 pKa = 5.6DD674 pKa = 4.7NFTQFSASVIKK685 pKa = 10.49FKK687 pKa = 11.11FKK689 pKa = 11.04ADD691 pKa = 3.78LVGSTSTFQFVANQINNISFEE712 pKa = 4.28HH713 pKa = 6.49YY714 pKa = 11.01NDD716 pKa = 3.24ISSNASTFNGNIGLTCFSLDD736 pKa = 3.26SDD738 pKa = 4.01GDD740 pKa = 4.14GIEE743 pKa = 5.74DD744 pKa = 4.25MFDD747 pKa = 4.28LDD749 pKa = 4.25TDD751 pKa = 3.61NDD753 pKa = 5.31GIPDD757 pKa = 4.28LYY759 pKa = 10.87DD760 pKa = 3.89AYY762 pKa = 10.64GQEE765 pKa = 3.76ISLAKK770 pKa = 9.87TDD772 pKa = 4.09ANLDD776 pKa = 3.52GLDD779 pKa = 4.41DD780 pKa = 3.94VFEE783 pKa = 5.33SITTNLDD790 pKa = 3.03TDD792 pKa = 3.88GDD794 pKa = 4.35GVNNYY799 pKa = 9.9LDD801 pKa = 4.0YY802 pKa = 11.49DD803 pKa = 3.92SDD805 pKa = 3.92NDD807 pKa = 4.57GIYY810 pKa = 10.82DD811 pKa = 3.66STEE814 pKa = 3.73ASHH817 pKa = 7.07GLDD820 pKa = 2.95TDD822 pKa = 4.07FNGTIDD828 pKa = 3.64TFIDD832 pKa = 3.22INKK835 pKa = 10.02NGLEE839 pKa = 4.24DD840 pKa = 3.7SLEE843 pKa = 4.22NDD845 pKa = 3.56TTVQTISLKK854 pKa = 9.4YY855 pKa = 9.17TIADD859 pKa = 3.57TDD861 pKa = 3.37ADD863 pKa = 4.26AIFNFIEE870 pKa = 4.69LDD872 pKa = 3.63SDD874 pKa = 4.25ADD876 pKa = 3.67GCNDD880 pKa = 3.29VIEE883 pKa = 5.78AGFTDD888 pKa = 3.86NNNDD892 pKa = 3.73GILFANSFSVDD903 pKa = 3.15EE904 pKa = 4.39NGKK907 pKa = 9.76VKK909 pKa = 10.76NPNDD913 pKa = 4.15GYY915 pKa = 7.67TTPNSNYY922 pKa = 6.83TTSAPIVVTTFTDD935 pKa = 3.44VTFCEE940 pKa = 4.22NLTDD944 pKa = 5.71IITIDD949 pKa = 3.56SNADD953 pKa = 3.06SFQWEE958 pKa = 4.49FSTDD962 pKa = 3.07GTNWLIINEE971 pKa = 4.62DD972 pKa = 3.93DD973 pKa = 4.24TTDD976 pKa = 3.89LYY978 pKa = 11.25QGSTSASLQITNTPFTYY995 pKa = 10.23DD996 pKa = 2.83SYY998 pKa = 11.32QYY1000 pKa = 10.66RR1001 pKa = 11.84VALNRR1006 pKa = 11.84TGNSCGFTSDD1016 pKa = 5.5AITLTVNPKK1025 pKa = 9.91PVIEE1029 pKa = 4.54NNPAALFQCDD1039 pKa = 4.04TDD1041 pKa = 4.67SDD1043 pKa = 4.14LQTTFNLTEE1052 pKa = 4.17SEE1054 pKa = 3.84ISITNEE1060 pKa = 3.12TDD1062 pKa = 3.32VNFEE1066 pKa = 4.23YY1067 pKa = 11.09YY1068 pKa = 9.36ATEE1071 pKa = 3.93LDD1073 pKa = 4.88AINGTPQVADD1083 pKa = 3.37KK1084 pKa = 10.72TSYY1087 pKa = 10.81SVNTNGEE1094 pKa = 4.05AWVKK1098 pKa = 9.78TISNTTACYY1107 pKa = 10.27SISKK1111 pKa = 10.27INLTVSYY1118 pKa = 10.76APNEE1122 pKa = 4.34TFNDD1126 pKa = 3.41TLYY1129 pKa = 10.97QCDD1132 pKa = 4.82DD1133 pKa = 4.27LLDD1136 pKa = 4.59LDD1138 pKa = 5.57GNNTTNNDD1146 pKa = 3.72DD1147 pKa = 3.69KK1148 pKa = 11.81DD1149 pKa = 4.54GITSFDD1155 pKa = 3.94LSNISDD1161 pKa = 4.48KK1162 pKa = 11.02ISTDD1166 pKa = 2.96SDD1168 pKa = 3.28IKK1170 pKa = 10.82IEE1172 pKa = 4.72FYY1174 pKa = 10.16EE1175 pKa = 4.22TDD1177 pKa = 3.48DD1178 pKa = 4.74DD1179 pKa = 4.03RR1180 pKa = 11.84TRR1182 pKa = 11.84SLNEE1186 pKa = 3.26ITEE1189 pKa = 4.42TQDD1192 pKa = 3.08LASYY1196 pKa = 10.37RR1197 pKa = 11.84NKK1199 pKa = 10.51NYY1201 pKa = 10.09PFTTGNPVTIFYY1213 pKa = 10.91KK1214 pKa = 10.8LISITNNNCQGIGEE1228 pKa = 4.97FYY1230 pKa = 11.2LEE1232 pKa = 4.24VNKK1235 pKa = 10.29IPEE1238 pKa = 4.06FTVDD1242 pKa = 4.24GEE1244 pKa = 4.44APSDD1248 pKa = 4.22PIIICAEE1255 pKa = 4.21NIPYY1259 pKa = 8.67TLNVYY1264 pKa = 8.93TSNSSYY1270 pKa = 11.06NYY1272 pKa = 9.35QWTNEE1277 pKa = 4.03KK1278 pKa = 10.56GDD1280 pKa = 3.97NLGTTQNINISDD1292 pKa = 3.67AGVYY1296 pKa = 8.72TVTSSTNSCSRR1307 pKa = 11.84SRR1309 pKa = 11.84TITVYY1314 pKa = 10.79KK1315 pKa = 10.5SDD1317 pKa = 3.87FEE1319 pKa = 4.22NLEE1322 pKa = 4.03EE1323 pKa = 4.45SFVTITDD1330 pKa = 3.69DD1331 pKa = 4.61LSVINSNLNIRR1342 pKa = 11.84IDD1344 pKa = 3.45IPTNPLINEE1353 pKa = 4.3VFQYY1357 pKa = 11.39ALEE1360 pKa = 5.29DD1361 pKa = 3.81EE1362 pKa = 5.24DD1363 pKa = 5.17GMTIRR1368 pKa = 11.84SFQDD1372 pKa = 3.03SNVFEE1377 pKa = 5.49DD1378 pKa = 3.98IEE1380 pKa = 4.41GGIYY1384 pKa = 9.98KK1385 pKa = 10.17IIVEE1389 pKa = 4.48SKK1391 pKa = 9.03NGCGTSEE1398 pKa = 4.47LLVSVIQFPKK1408 pKa = 10.42FFTPNGDD1415 pKa = 3.34RR1416 pKa = 11.84KK1417 pKa = 10.33NDD1419 pKa = 2.79SWGIKK1424 pKa = 9.47GVSNTLYY1431 pKa = 10.49QSNSSINIFNRR1442 pKa = 11.84YY1443 pKa = 7.94GKK1445 pKa = 10.34LVAQTTIDD1453 pKa = 3.51GDD1455 pKa = 3.88GWDD1458 pKa = 3.44GTYY1461 pKa = 10.52NGKK1464 pKa = 9.2LLPSDD1469 pKa = 4.78DD1470 pKa = 3.67YY1471 pKa = 10.8WYY1473 pKa = 10.48SIQLIPVNTDD1483 pKa = 2.8QKK1485 pKa = 10.67PILKK1489 pKa = 9.71KK1490 pKa = 10.6GHH1492 pKa = 6.37FSLIRR1497 pKa = 11.84KK1498 pKa = 8.22
Molecular weight: 164.3 kDa
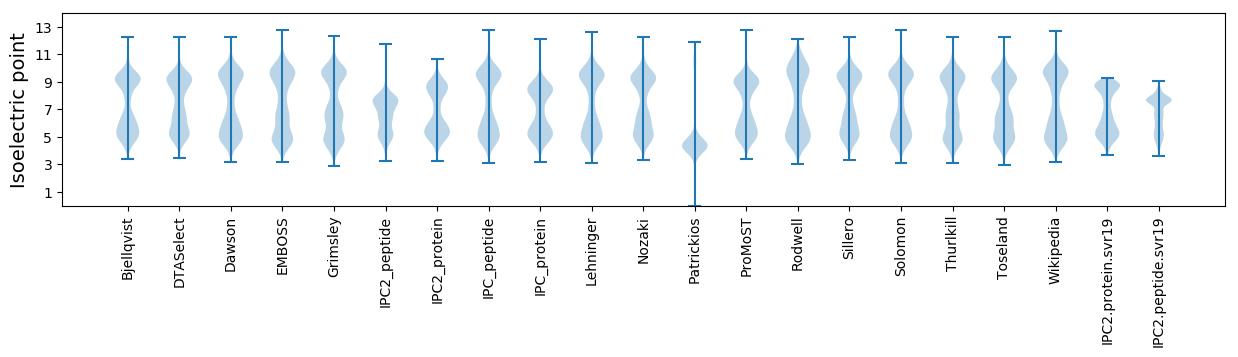
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A176TAX6|A0A176TAX6_9FLAO Glutamate-1-semialdehyde 2 1-aminomutase OS=Polaribacter atrinae OX=1333662 GN=hemL PE=3 SV=1
MM1 pKa = 7.51ARR3 pKa = 11.84SLKK6 pKa = 10.37LWLSNTEE13 pKa = 3.61AEE15 pKa = 4.24KK16 pKa = 10.87YY17 pKa = 10.09IRR19 pKa = 11.84VSKK22 pKa = 8.47TLKK25 pKa = 9.92PYY27 pKa = 10.31IMKK30 pKa = 10.36KK31 pKa = 10.36GVLILIGMFFLVSTIKK47 pKa = 10.91AEE49 pKa = 4.21NNKK52 pKa = 9.29EE53 pKa = 4.1VPNKK57 pKa = 10.13IGFNYY62 pKa = 10.23SYY64 pKa = 10.62EE65 pKa = 4.08NTVNFVEE72 pKa = 4.4RR73 pKa = 11.84DD74 pKa = 3.33VEE76 pKa = 4.92FFIFTNGEE84 pKa = 3.72FDD86 pKa = 4.53FNTHH90 pKa = 4.44YY91 pKa = 10.05TSNRR95 pKa = 11.84NEE97 pKa = 3.59IGVFISRR104 pKa = 11.84DD105 pKa = 3.17YY106 pKa = 10.57RR107 pKa = 11.84GRR109 pKa = 11.84IEE111 pKa = 5.14RR112 pKa = 11.84IGNSFINYY120 pKa = 8.99DD121 pKa = 3.08RR122 pKa = 11.84YY123 pKa = 11.52GNVTRR128 pKa = 11.84IGNVFMKK135 pKa = 9.87YY136 pKa = 9.52YY137 pKa = 10.35RR138 pKa = 11.84GQLTDD143 pKa = 3.25VGNLKK148 pKa = 9.66IRR150 pKa = 11.84YY151 pKa = 7.53DD152 pKa = 2.91HH153 pKa = 6.7WGNPKK158 pKa = 10.17FYY160 pKa = 11.29GNVKK164 pKa = 10.41DD165 pKa = 5.26NYY167 pKa = 8.65YY168 pKa = 10.23HH169 pKa = 6.91YY170 pKa = 11.24NGLRR174 pKa = 11.84INLNIGNIFEE184 pKa = 4.66YY185 pKa = 10.38NNSYY189 pKa = 10.36FSHH192 pKa = 7.28RR193 pKa = 11.84DD194 pKa = 3.41FARR197 pKa = 11.84HH198 pKa = 4.36YY199 pKa = 11.1SKK201 pKa = 10.38IRR203 pKa = 11.84EE204 pKa = 4.14DD205 pKa = 3.2NKK207 pKa = 10.1YY208 pKa = 10.49YY209 pKa = 10.58YY210 pKa = 11.05YY211 pKa = 10.92KK212 pKa = 10.81ANPNAKK218 pKa = 9.29IGNKK222 pKa = 9.81SKK224 pKa = 10.43ILRR227 pKa = 11.84RR228 pKa = 11.84RR229 pKa = 11.84KK230 pKa = 8.52PAPVTRR236 pKa = 11.84RR237 pKa = 11.84SNTTYY242 pKa = 10.61RR243 pKa = 11.84KK244 pKa = 9.41PVTINSNRR252 pKa = 11.84RR253 pKa = 11.84TVVGKK258 pKa = 10.49RR259 pKa = 11.84NNSTTRR265 pKa = 11.84RR266 pKa = 11.84STTINTNKK274 pKa = 10.27NGTIDD279 pKa = 3.38RR280 pKa = 11.84RR281 pKa = 11.84NNSTVTRR288 pKa = 11.84RR289 pKa = 11.84PTTVTRR295 pKa = 11.84GKK297 pKa = 6.52TTSVKK302 pKa = 10.76RR303 pKa = 11.84NDD305 pKa = 3.4NNSNRR310 pKa = 11.84KK311 pKa = 6.39PTVVKK316 pKa = 10.53RR317 pKa = 11.84KK318 pKa = 9.14NEE320 pKa = 4.05KK321 pKa = 10.21KK322 pKa = 10.29KK323 pKa = 10.27ATTKK327 pKa = 10.58EE328 pKa = 3.67EE329 pKa = 4.09NNRR332 pKa = 11.84KK333 pKa = 9.23RR334 pKa = 11.84RR335 pKa = 11.84SS336 pKa = 3.28
MM1 pKa = 7.51ARR3 pKa = 11.84SLKK6 pKa = 10.37LWLSNTEE13 pKa = 3.61AEE15 pKa = 4.24KK16 pKa = 10.87YY17 pKa = 10.09IRR19 pKa = 11.84VSKK22 pKa = 8.47TLKK25 pKa = 9.92PYY27 pKa = 10.31IMKK30 pKa = 10.36KK31 pKa = 10.36GVLILIGMFFLVSTIKK47 pKa = 10.91AEE49 pKa = 4.21NNKK52 pKa = 9.29EE53 pKa = 4.1VPNKK57 pKa = 10.13IGFNYY62 pKa = 10.23SYY64 pKa = 10.62EE65 pKa = 4.08NTVNFVEE72 pKa = 4.4RR73 pKa = 11.84DD74 pKa = 3.33VEE76 pKa = 4.92FFIFTNGEE84 pKa = 3.72FDD86 pKa = 4.53FNTHH90 pKa = 4.44YY91 pKa = 10.05TSNRR95 pKa = 11.84NEE97 pKa = 3.59IGVFISRR104 pKa = 11.84DD105 pKa = 3.17YY106 pKa = 10.57RR107 pKa = 11.84GRR109 pKa = 11.84IEE111 pKa = 5.14RR112 pKa = 11.84IGNSFINYY120 pKa = 8.99DD121 pKa = 3.08RR122 pKa = 11.84YY123 pKa = 11.52GNVTRR128 pKa = 11.84IGNVFMKK135 pKa = 9.87YY136 pKa = 9.52YY137 pKa = 10.35RR138 pKa = 11.84GQLTDD143 pKa = 3.25VGNLKK148 pKa = 9.66IRR150 pKa = 11.84YY151 pKa = 7.53DD152 pKa = 2.91HH153 pKa = 6.7WGNPKK158 pKa = 10.17FYY160 pKa = 11.29GNVKK164 pKa = 10.41DD165 pKa = 5.26NYY167 pKa = 8.65YY168 pKa = 10.23HH169 pKa = 6.91YY170 pKa = 11.24NGLRR174 pKa = 11.84INLNIGNIFEE184 pKa = 4.66YY185 pKa = 10.38NNSYY189 pKa = 10.36FSHH192 pKa = 7.28RR193 pKa = 11.84DD194 pKa = 3.41FARR197 pKa = 11.84HH198 pKa = 4.36YY199 pKa = 11.1SKK201 pKa = 10.38IRR203 pKa = 11.84EE204 pKa = 4.14DD205 pKa = 3.2NKK207 pKa = 10.1YY208 pKa = 10.49YY209 pKa = 10.58YY210 pKa = 11.05YY211 pKa = 10.92KK212 pKa = 10.81ANPNAKK218 pKa = 9.29IGNKK222 pKa = 9.81SKK224 pKa = 10.43ILRR227 pKa = 11.84RR228 pKa = 11.84RR229 pKa = 11.84KK230 pKa = 8.52PAPVTRR236 pKa = 11.84RR237 pKa = 11.84SNTTYY242 pKa = 10.61RR243 pKa = 11.84KK244 pKa = 9.41PVTINSNRR252 pKa = 11.84RR253 pKa = 11.84TVVGKK258 pKa = 10.49RR259 pKa = 11.84NNSTTRR265 pKa = 11.84RR266 pKa = 11.84STTINTNKK274 pKa = 10.27NGTIDD279 pKa = 3.38RR280 pKa = 11.84RR281 pKa = 11.84NNSTVTRR288 pKa = 11.84RR289 pKa = 11.84PTTVTRR295 pKa = 11.84GKK297 pKa = 6.52TTSVKK302 pKa = 10.76RR303 pKa = 11.84NDD305 pKa = 3.4NNSNRR310 pKa = 11.84KK311 pKa = 6.39PTVVKK316 pKa = 10.53RR317 pKa = 11.84KK318 pKa = 9.14NEE320 pKa = 4.05KK321 pKa = 10.21KK322 pKa = 10.29KK323 pKa = 10.27ATTKK327 pKa = 10.58EE328 pKa = 3.67EE329 pKa = 4.09NNRR332 pKa = 11.84KK333 pKa = 9.23RR334 pKa = 11.84RR335 pKa = 11.84SS336 pKa = 3.28
Molecular weight: 39.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1114503 |
50 |
2354 |
342.1 |
38.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.044 ± 0.039 | 0.666 ± 0.012 |
5.535 ± 0.034 | 6.685 ± 0.036 |
5.43 ± 0.036 | 6.143 ± 0.043 |
1.641 ± 0.018 | 8.348 ± 0.043 |
8.665 ± 0.055 | 9.183 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.002 ± 0.021 | 6.592 ± 0.045 |
3.081 ± 0.021 | 3.121 ± 0.023 |
3.072 ± 0.027 | 6.595 ± 0.035 |
5.998 ± 0.04 | 6.156 ± 0.031 |
1.02 ± 0.015 | 4.025 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
