
Equus caballus papillomavirus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyoiotapapillomavirus; Dyoiotapapillomavirus 2
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
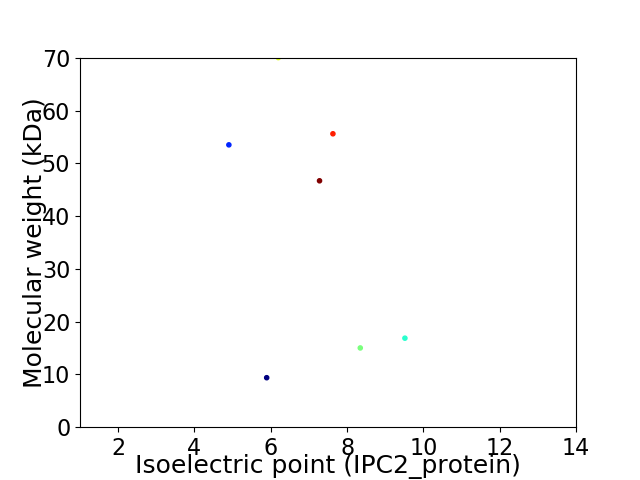
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K9M9L3|K9M9L3_9PAPI Replication protein E1 OS=Equus caballus papillomavirus 4 OX=1235428 GN=E1 PE=3 SV=1
MM1 pKa = 7.49EE2 pKa = 5.26AQPPLKK8 pKa = 9.7RR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.12RR12 pKa = 11.84ASVEE16 pKa = 3.41NLYY19 pKa = 10.55RR20 pKa = 11.84SCKK23 pKa = 10.44LGGDD27 pKa = 4.01CPADD31 pKa = 3.24VVPKK35 pKa = 10.92VEE37 pKa = 4.48GDD39 pKa = 3.6TPADD43 pKa = 5.26RR44 pKa = 11.84ILKK47 pKa = 8.66WLSSFLFFGNLGISSTGRR65 pKa = 11.84GVGGFRR71 pKa = 11.84PLGGGAHH78 pKa = 6.3AAEE81 pKa = 5.02GGSVRR86 pKa = 11.84VGTPISTVEE95 pKa = 3.9VGSAVDD101 pKa = 3.55SVGPLEE107 pKa = 4.49LAPIDD112 pKa = 4.07GVTPTSPAVVSMGEE126 pKa = 4.32GVGEE130 pKa = 4.06AVDD133 pKa = 3.74VFAEE137 pKa = 4.22VTPPVGGATIGSEE150 pKa = 4.25VNSAGVGEE158 pKa = 4.11PAILDD163 pKa = 3.4VSAEE167 pKa = 4.2VVPKK171 pKa = 10.42LRR173 pKa = 11.84AAVTSTEE180 pKa = 3.77FHH182 pKa = 6.19NPAFQVEE189 pKa = 4.57FRR191 pKa = 11.84SVGTGEE197 pKa = 4.24TSGSEE202 pKa = 3.52QAYY205 pKa = 8.01WFGHH209 pKa = 5.93AGGHH213 pKa = 5.53AVGEE217 pKa = 4.22AGEE220 pKa = 4.48SIEE223 pKa = 5.84LVDD226 pKa = 5.09LNYY229 pKa = 10.53RR230 pKa = 11.84GPKK233 pKa = 9.02TSTPSAEE240 pKa = 4.37VVSEE244 pKa = 3.52PRR246 pKa = 11.84LWGRR250 pKa = 11.84RR251 pKa = 11.84FQQVRR256 pKa = 11.84VQQPEE261 pKa = 4.5FVTAPQTLVEE271 pKa = 4.94FGFSNPAYY279 pKa = 10.08DD280 pKa = 4.18PEE282 pKa = 6.22GSLLFTYY289 pKa = 10.72NEE291 pKa = 4.41SAPSAAPSSPFQDD304 pKa = 2.45IAYY307 pKa = 9.85LGRR310 pKa = 11.84PEE312 pKa = 3.38ITARR316 pKa = 11.84AGRR319 pKa = 11.84VGVSRR324 pKa = 11.84YY325 pKa = 8.26GRR327 pKa = 11.84RR328 pKa = 11.84ATLHH332 pKa = 5.37TRR334 pKa = 11.84AGTILGGEE342 pKa = 3.88VHH344 pKa = 6.39FRR346 pKa = 11.84YY347 pKa = 10.13DD348 pKa = 3.62FSSIEE353 pKa = 4.11PEE355 pKa = 4.03TLEE358 pKa = 5.51LSDD361 pKa = 5.0LTVQDD366 pKa = 3.86STSLAGLSSVDD377 pKa = 3.81LGSEE381 pKa = 3.97GMSINEE387 pKa = 3.78SWLGEE392 pKa = 3.98DD393 pKa = 3.86EE394 pKa = 4.18QVSFHH399 pKa = 6.28GVLAFSEE406 pKa = 4.72GSEE409 pKa = 4.38AEE411 pKa = 3.81IAPYY415 pKa = 9.29PRR417 pKa = 11.84SVWVRR422 pKa = 11.84LAGEE426 pKa = 4.48GSSFSFWGGEE436 pKa = 3.99NLPGRR441 pKa = 11.84AVFITSLPSPSATPPTSTDD460 pKa = 2.98TAGFSMLDD468 pKa = 3.19PSLLKK473 pKa = 9.83RR474 pKa = 11.84CRR476 pKa = 11.84RR477 pKa = 11.84KK478 pKa = 9.99RR479 pKa = 11.84RR480 pKa = 11.84SCFADD485 pKa = 4.58GIVDD489 pKa = 3.89ARR491 pKa = 11.84QSEE494 pKa = 4.94VLSAPCSCDD503 pKa = 3.8KK504 pKa = 10.78GAQYY508 pKa = 11.55
MM1 pKa = 7.49EE2 pKa = 5.26AQPPLKK8 pKa = 9.7RR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.12RR12 pKa = 11.84ASVEE16 pKa = 3.41NLYY19 pKa = 10.55RR20 pKa = 11.84SCKK23 pKa = 10.44LGGDD27 pKa = 4.01CPADD31 pKa = 3.24VVPKK35 pKa = 10.92VEE37 pKa = 4.48GDD39 pKa = 3.6TPADD43 pKa = 5.26RR44 pKa = 11.84ILKK47 pKa = 8.66WLSSFLFFGNLGISSTGRR65 pKa = 11.84GVGGFRR71 pKa = 11.84PLGGGAHH78 pKa = 6.3AAEE81 pKa = 5.02GGSVRR86 pKa = 11.84VGTPISTVEE95 pKa = 3.9VGSAVDD101 pKa = 3.55SVGPLEE107 pKa = 4.49LAPIDD112 pKa = 4.07GVTPTSPAVVSMGEE126 pKa = 4.32GVGEE130 pKa = 4.06AVDD133 pKa = 3.74VFAEE137 pKa = 4.22VTPPVGGATIGSEE150 pKa = 4.25VNSAGVGEE158 pKa = 4.11PAILDD163 pKa = 3.4VSAEE167 pKa = 4.2VVPKK171 pKa = 10.42LRR173 pKa = 11.84AAVTSTEE180 pKa = 3.77FHH182 pKa = 6.19NPAFQVEE189 pKa = 4.57FRR191 pKa = 11.84SVGTGEE197 pKa = 4.24TSGSEE202 pKa = 3.52QAYY205 pKa = 8.01WFGHH209 pKa = 5.93AGGHH213 pKa = 5.53AVGEE217 pKa = 4.22AGEE220 pKa = 4.48SIEE223 pKa = 5.84LVDD226 pKa = 5.09LNYY229 pKa = 10.53RR230 pKa = 11.84GPKK233 pKa = 9.02TSTPSAEE240 pKa = 4.37VVSEE244 pKa = 3.52PRR246 pKa = 11.84LWGRR250 pKa = 11.84RR251 pKa = 11.84FQQVRR256 pKa = 11.84VQQPEE261 pKa = 4.5FVTAPQTLVEE271 pKa = 4.94FGFSNPAYY279 pKa = 10.08DD280 pKa = 4.18PEE282 pKa = 6.22GSLLFTYY289 pKa = 10.72NEE291 pKa = 4.41SAPSAAPSSPFQDD304 pKa = 2.45IAYY307 pKa = 9.85LGRR310 pKa = 11.84PEE312 pKa = 3.38ITARR316 pKa = 11.84AGRR319 pKa = 11.84VGVSRR324 pKa = 11.84YY325 pKa = 8.26GRR327 pKa = 11.84RR328 pKa = 11.84ATLHH332 pKa = 5.37TRR334 pKa = 11.84AGTILGGEE342 pKa = 3.88VHH344 pKa = 6.39FRR346 pKa = 11.84YY347 pKa = 10.13DD348 pKa = 3.62FSSIEE353 pKa = 4.11PEE355 pKa = 4.03TLEE358 pKa = 5.51LSDD361 pKa = 5.0LTVQDD366 pKa = 3.86STSLAGLSSVDD377 pKa = 3.81LGSEE381 pKa = 3.97GMSINEE387 pKa = 3.78SWLGEE392 pKa = 3.98DD393 pKa = 3.86EE394 pKa = 4.18QVSFHH399 pKa = 6.28GVLAFSEE406 pKa = 4.72GSEE409 pKa = 4.38AEE411 pKa = 3.81IAPYY415 pKa = 9.29PRR417 pKa = 11.84SVWVRR422 pKa = 11.84LAGEE426 pKa = 4.48GSSFSFWGGEE436 pKa = 3.99NLPGRR441 pKa = 11.84AVFITSLPSPSATPPTSTDD460 pKa = 2.98TAGFSMLDD468 pKa = 3.19PSLLKK473 pKa = 9.83RR474 pKa = 11.84CRR476 pKa = 11.84RR477 pKa = 11.84KK478 pKa = 9.99RR479 pKa = 11.84RR480 pKa = 11.84SCFADD485 pKa = 4.58GIVDD489 pKa = 3.89ARR491 pKa = 11.84QSEE494 pKa = 4.94VLSAPCSCDD503 pKa = 3.8KK504 pKa = 10.78GAQYY508 pKa = 11.55
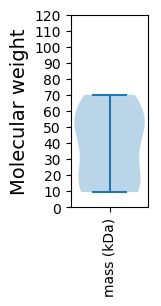
Molecular weight: 53.51 kDa
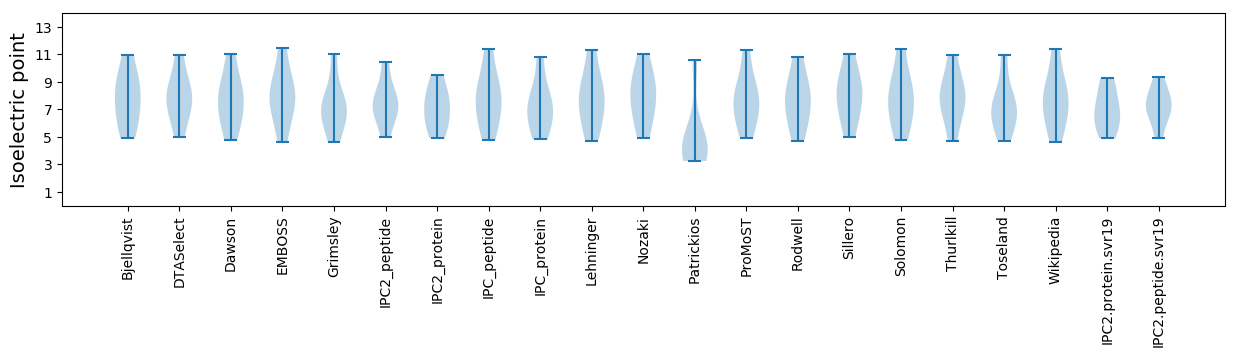
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K9M9S8|K9M9S8_9PAPI E4 OS=Equus caballus papillomavirus 4 OX=1235428 PE=4 SV=1
MM1 pKa = 7.69KK2 pKa = 9.39LTSTLLLAAPPTEE15 pKa = 4.38TPRR18 pKa = 11.84TPQTEE23 pKa = 4.37LQGGPPPPLDD33 pKa = 4.17LQPGPQSPAVQLPEE47 pKa = 3.96TLQQFLRR54 pKa = 11.84NSHH57 pKa = 5.36IWGGDD62 pKa = 2.97VWPRR66 pKa = 11.84RR67 pKa = 11.84GGGYY71 pKa = 7.9TRR73 pKa = 11.84QQGTSLLSTQAACLGHH89 pKa = 6.58SGAPPCKK96 pKa = 9.47PGLDD100 pKa = 3.69GGASGEE106 pKa = 4.37WNLPPPPAALRR117 pKa = 11.84ARR119 pKa = 11.84LRR121 pKa = 11.84QTRR124 pKa = 11.84LRR126 pKa = 11.84NRR128 pKa = 11.84RR129 pKa = 11.84RR130 pKa = 11.84LRR132 pKa = 11.84RR133 pKa = 11.84CLEE136 pKa = 3.96LLRR139 pKa = 11.84ADD141 pKa = 4.24LEE143 pKa = 4.22EE144 pKa = 3.91LRR146 pKa = 11.84FWVTVGLLL154 pKa = 3.32
MM1 pKa = 7.69KK2 pKa = 9.39LTSTLLLAAPPTEE15 pKa = 4.38TPRR18 pKa = 11.84TPQTEE23 pKa = 4.37LQGGPPPPLDD33 pKa = 4.17LQPGPQSPAVQLPEE47 pKa = 3.96TLQQFLRR54 pKa = 11.84NSHH57 pKa = 5.36IWGGDD62 pKa = 2.97VWPRR66 pKa = 11.84RR67 pKa = 11.84GGGYY71 pKa = 7.9TRR73 pKa = 11.84QQGTSLLSTQAACLGHH89 pKa = 6.58SGAPPCKK96 pKa = 9.47PGLDD100 pKa = 3.69GGASGEE106 pKa = 4.37WNLPPPPAALRR117 pKa = 11.84ARR119 pKa = 11.84LRR121 pKa = 11.84QTRR124 pKa = 11.84LRR126 pKa = 11.84NRR128 pKa = 11.84RR129 pKa = 11.84RR130 pKa = 11.84LRR132 pKa = 11.84RR133 pKa = 11.84CLEE136 pKa = 3.96LLRR139 pKa = 11.84ADD141 pKa = 4.24LEE143 pKa = 4.22EE144 pKa = 3.91LRR146 pKa = 11.84FWVTVGLLL154 pKa = 3.32
Molecular weight: 16.87 kDa
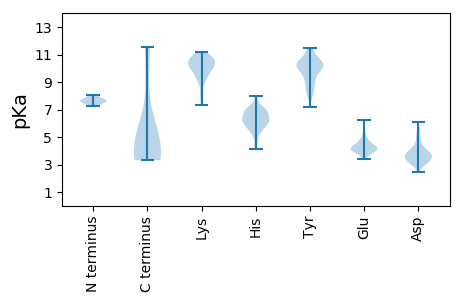
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2425 |
85 |
626 |
346.4 |
38.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.742 ± 0.605 | 2.227 ± 0.667 |
4.742 ± 0.4 | 6.103 ± 0.499 |
4.371 ± 0.605 | 8.619 ± 1.133 |
1.979 ± 0.271 | 2.598 ± 0.369 |
3.629 ± 0.684 | 8.784 ± 0.827 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.567 ± 0.259 | 3.216 ± 0.777 |
7.216 ± 1.009 | 4.206 ± 0.514 |
7.258 ± 0.607 | 7.67 ± 1.028 |
5.773 ± 0.334 | 6.515 ± 0.797 |
1.856 ± 0.232 | 2.928 ± 0.621 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
