
Hubei tombus-like virus 38
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.16
Get precalculated fractions of proteins
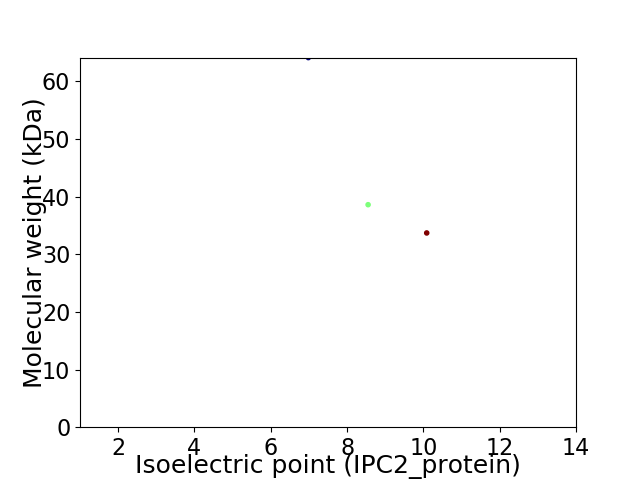
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGW0|A0A1L3KGW0_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 38 OX=1923286 PE=4 SV=1
MM1 pKa = 7.7SYY3 pKa = 10.74NVSEE7 pKa = 4.41VHH9 pKa = 6.14LQPLTRR15 pKa = 11.84ARR17 pKa = 11.84VSRR20 pKa = 11.84PFKK23 pKa = 10.52SRR25 pKa = 11.84GWYY28 pKa = 10.09LMPSILIPEE37 pKa = 4.18QTAMVPMVDD46 pKa = 4.83LEE48 pKa = 4.19NATASLATRR57 pKa = 11.84VVVLEE62 pKa = 4.24PKK64 pKa = 10.61QIDD67 pKa = 3.63FNLAKK72 pKa = 10.58LPEE75 pKa = 4.12TRR77 pKa = 11.84AKK79 pKa = 10.21YY80 pKa = 10.58LRR82 pKa = 11.84TKK84 pKa = 10.39CQFEE88 pKa = 4.25TRR90 pKa = 11.84ITAAEE95 pKa = 3.69AAAIEE100 pKa = 4.45EE101 pKa = 4.49EE102 pKa = 4.71VANDD106 pKa = 3.95PDD108 pKa = 3.75RR109 pKa = 11.84QLPYY113 pKa = 10.25QGRR116 pKa = 11.84LRR118 pKa = 11.84KK119 pKa = 9.45LVAEE123 pKa = 4.93FVRR126 pKa = 11.84DD127 pKa = 3.66PRR129 pKa = 11.84TQLPTLIQPLPLEE142 pKa = 4.62EE143 pKa = 4.18YY144 pKa = 7.71VTRR147 pKa = 11.84YY148 pKa = 9.89KK149 pKa = 10.57PAEE152 pKa = 3.79QRR154 pKa = 11.84HH155 pKa = 6.04LIEE158 pKa = 4.06EE159 pKa = 4.22AAKK162 pKa = 10.49VRR164 pKa = 11.84ATFVANSSVKK174 pKa = 10.72AFIKK178 pKa = 10.65QEE180 pKa = 4.1AGPPGDD186 pKa = 4.38PRR188 pKa = 11.84NISPRR193 pKa = 11.84SPGYY197 pKa = 9.47KK198 pKa = 9.79ACLGPYY204 pKa = 9.39IAAFEE209 pKa = 4.08AAMKK213 pKa = 9.89NCSYY217 pKa = 10.58LIKK220 pKa = 10.73GLTLPEE226 pKa = 4.32RR227 pKa = 11.84NVKK230 pKa = 9.34MSSIMMGLYY239 pKa = 10.17DD240 pKa = 4.91GSVIDD245 pKa = 4.02IDD247 pKa = 4.24FSRR250 pKa = 11.84FDD252 pKa = 3.14MHH254 pKa = 6.67IQHH257 pKa = 7.18DD258 pKa = 3.97VLLYY262 pKa = 10.27IEE264 pKa = 4.91HH265 pKa = 7.05EE266 pKa = 4.6CIKK269 pKa = 10.59HH270 pKa = 5.56AFGYY274 pKa = 10.83DD275 pKa = 3.65PFLCWLLDD283 pKa = 3.57MQLVTKK289 pKa = 9.95GYY291 pKa = 8.89HH292 pKa = 4.61VCGVAYY298 pKa = 7.54HH299 pKa = 6.14TKK301 pKa = 10.41GGRR304 pKa = 11.84CSGDD308 pKa = 3.16VNTSIGNALINRR320 pKa = 11.84FCSWYY325 pKa = 9.92SARR328 pKa = 11.84TLKK331 pKa = 11.05LGDD334 pKa = 3.18NWEE337 pKa = 4.47SFHH340 pKa = 7.27EE341 pKa = 4.79GDD343 pKa = 5.3DD344 pKa = 4.3TIWWCSGLLVRR355 pKa = 11.84PNWIMYY361 pKa = 9.01VVEE364 pKa = 4.44LCHH367 pKa = 6.8LRR369 pKa = 11.84LGFPISISLNRR380 pKa = 11.84TLDD383 pKa = 3.47TVDD386 pKa = 3.5FCGRR390 pKa = 11.84VFYY393 pKa = 10.0TLQDD397 pKa = 3.42KK398 pKa = 9.0VMCMADD404 pKa = 3.54PVRR407 pKa = 11.84TLQKK411 pKa = 10.16FCITSRR417 pKa = 11.84PVQRR421 pKa = 11.84DD422 pKa = 3.4QYY424 pKa = 10.28SPTVAKK430 pKa = 10.39EE431 pKa = 3.82LLLAKK436 pKa = 10.56AMAYY440 pKa = 10.32FSTDD444 pKa = 2.45RR445 pKa = 11.84QMPIIGPLCWWIIQLLKK462 pKa = 10.69FEE464 pKa = 4.31EE465 pKa = 4.56HH466 pKa = 4.92VVPRR470 pKa = 11.84FEE472 pKa = 6.04HH473 pKa = 6.16DD474 pKa = 3.43TQFRR478 pKa = 11.84ADD480 pKa = 3.51LAGVVSKK487 pKa = 10.02YY488 pKa = 6.83EE489 pKa = 3.68WRR491 pKa = 11.84QFPTIPGEE499 pKa = 3.76ARR501 pKa = 11.84MLFFMRR507 pKa = 11.84TGIYY511 pKa = 9.61PQEE514 pKa = 3.8QVLWEE519 pKa = 4.14RR520 pKa = 11.84VVVTLPFIPEE530 pKa = 3.77VMPQLRR536 pKa = 11.84LAQIKK541 pKa = 10.77DD542 pKa = 3.86FDD544 pKa = 4.71PLRR547 pKa = 11.84HH548 pKa = 5.63MPIDD552 pKa = 3.66GYY554 pKa = 10.12MFEE557 pKa = 4.48
MM1 pKa = 7.7SYY3 pKa = 10.74NVSEE7 pKa = 4.41VHH9 pKa = 6.14LQPLTRR15 pKa = 11.84ARR17 pKa = 11.84VSRR20 pKa = 11.84PFKK23 pKa = 10.52SRR25 pKa = 11.84GWYY28 pKa = 10.09LMPSILIPEE37 pKa = 4.18QTAMVPMVDD46 pKa = 4.83LEE48 pKa = 4.19NATASLATRR57 pKa = 11.84VVVLEE62 pKa = 4.24PKK64 pKa = 10.61QIDD67 pKa = 3.63FNLAKK72 pKa = 10.58LPEE75 pKa = 4.12TRR77 pKa = 11.84AKK79 pKa = 10.21YY80 pKa = 10.58LRR82 pKa = 11.84TKK84 pKa = 10.39CQFEE88 pKa = 4.25TRR90 pKa = 11.84ITAAEE95 pKa = 3.69AAAIEE100 pKa = 4.45EE101 pKa = 4.49EE102 pKa = 4.71VANDD106 pKa = 3.95PDD108 pKa = 3.75RR109 pKa = 11.84QLPYY113 pKa = 10.25QGRR116 pKa = 11.84LRR118 pKa = 11.84KK119 pKa = 9.45LVAEE123 pKa = 4.93FVRR126 pKa = 11.84DD127 pKa = 3.66PRR129 pKa = 11.84TQLPTLIQPLPLEE142 pKa = 4.62EE143 pKa = 4.18YY144 pKa = 7.71VTRR147 pKa = 11.84YY148 pKa = 9.89KK149 pKa = 10.57PAEE152 pKa = 3.79QRR154 pKa = 11.84HH155 pKa = 6.04LIEE158 pKa = 4.06EE159 pKa = 4.22AAKK162 pKa = 10.49VRR164 pKa = 11.84ATFVANSSVKK174 pKa = 10.72AFIKK178 pKa = 10.65QEE180 pKa = 4.1AGPPGDD186 pKa = 4.38PRR188 pKa = 11.84NISPRR193 pKa = 11.84SPGYY197 pKa = 9.47KK198 pKa = 9.79ACLGPYY204 pKa = 9.39IAAFEE209 pKa = 4.08AAMKK213 pKa = 9.89NCSYY217 pKa = 10.58LIKK220 pKa = 10.73GLTLPEE226 pKa = 4.32RR227 pKa = 11.84NVKK230 pKa = 9.34MSSIMMGLYY239 pKa = 10.17DD240 pKa = 4.91GSVIDD245 pKa = 4.02IDD247 pKa = 4.24FSRR250 pKa = 11.84FDD252 pKa = 3.14MHH254 pKa = 6.67IQHH257 pKa = 7.18DD258 pKa = 3.97VLLYY262 pKa = 10.27IEE264 pKa = 4.91HH265 pKa = 7.05EE266 pKa = 4.6CIKK269 pKa = 10.59HH270 pKa = 5.56AFGYY274 pKa = 10.83DD275 pKa = 3.65PFLCWLLDD283 pKa = 3.57MQLVTKK289 pKa = 9.95GYY291 pKa = 8.89HH292 pKa = 4.61VCGVAYY298 pKa = 7.54HH299 pKa = 6.14TKK301 pKa = 10.41GGRR304 pKa = 11.84CSGDD308 pKa = 3.16VNTSIGNALINRR320 pKa = 11.84FCSWYY325 pKa = 9.92SARR328 pKa = 11.84TLKK331 pKa = 11.05LGDD334 pKa = 3.18NWEE337 pKa = 4.47SFHH340 pKa = 7.27EE341 pKa = 4.79GDD343 pKa = 5.3DD344 pKa = 4.3TIWWCSGLLVRR355 pKa = 11.84PNWIMYY361 pKa = 9.01VVEE364 pKa = 4.44LCHH367 pKa = 6.8LRR369 pKa = 11.84LGFPISISLNRR380 pKa = 11.84TLDD383 pKa = 3.47TVDD386 pKa = 3.5FCGRR390 pKa = 11.84VFYY393 pKa = 10.0TLQDD397 pKa = 3.42KK398 pKa = 9.0VMCMADD404 pKa = 3.54PVRR407 pKa = 11.84TLQKK411 pKa = 10.16FCITSRR417 pKa = 11.84PVQRR421 pKa = 11.84DD422 pKa = 3.4QYY424 pKa = 10.28SPTVAKK430 pKa = 10.39EE431 pKa = 3.82LLLAKK436 pKa = 10.56AMAYY440 pKa = 10.32FSTDD444 pKa = 2.45RR445 pKa = 11.84QMPIIGPLCWWIIQLLKK462 pKa = 10.69FEE464 pKa = 4.31EE465 pKa = 4.56HH466 pKa = 4.92VVPRR470 pKa = 11.84FEE472 pKa = 6.04HH473 pKa = 6.16DD474 pKa = 3.43TQFRR478 pKa = 11.84ADD480 pKa = 3.51LAGVVSKK487 pKa = 10.02YY488 pKa = 6.83EE489 pKa = 3.68WRR491 pKa = 11.84QFPTIPGEE499 pKa = 3.76ARR501 pKa = 11.84MLFFMRR507 pKa = 11.84TGIYY511 pKa = 9.61PQEE514 pKa = 3.8QVLWEE519 pKa = 4.14RR520 pKa = 11.84VVVTLPFIPEE530 pKa = 3.77VMPQLRR536 pKa = 11.84LAQIKK541 pKa = 10.77DD542 pKa = 3.86FDD544 pKa = 4.71PLRR547 pKa = 11.84HH548 pKa = 5.63MPIDD552 pKa = 3.66GYY554 pKa = 10.12MFEE557 pKa = 4.48
Molecular weight: 64.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGW0|A0A1L3KGW0_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 38 OX=1923286 PE=4 SV=1
MM1 pKa = 7.31EE2 pKa = 5.28AATLVVADD10 pKa = 4.08NQMDD14 pKa = 3.92RR15 pKa = 11.84NKK17 pKa = 9.95TYY19 pKa = 11.21ARR21 pKa = 11.84VVQILRR27 pKa = 11.84SRR29 pKa = 11.84MASRR33 pKa = 11.84QADD36 pKa = 3.52MTILSEE42 pKa = 4.19VATITLRR49 pKa = 11.84LADD52 pKa = 4.76NINRR56 pKa = 11.84DD57 pKa = 3.54AVLSGNFRR65 pKa = 11.84GTILDD70 pKa = 3.67YY71 pKa = 10.53FDD73 pKa = 3.39RR74 pKa = 11.84RR75 pKa = 11.84GWVAYY80 pKa = 9.15YY81 pKa = 10.35LIAPLGTWLPRR92 pKa = 11.84SVWGWMLSKK101 pKa = 8.33MTSRR105 pKa = 11.84EE106 pKa = 3.62HH107 pKa = 7.19RR108 pKa = 11.84FNRR111 pKa = 11.84WVWPHH116 pKa = 5.83VSATHH121 pKa = 6.92CSRR124 pKa = 11.84IMSADD129 pKa = 3.06EE130 pKa = 3.79LQRR133 pKa = 11.84IRR135 pKa = 11.84GALTAIDD142 pKa = 3.93PGARR146 pKa = 11.84QPPFQEE152 pKa = 4.49PGLVLNAIHH161 pKa = 7.11SDD163 pKa = 3.35PGTDD167 pKa = 3.17SDD169 pKa = 4.27GTHH172 pKa = 5.57GRR174 pKa = 11.84PRR176 pKa = 11.84EE177 pKa = 3.83RR178 pKa = 11.84DD179 pKa = 3.56RR180 pKa = 11.84IPSNPSSSTRR190 pKa = 11.84AEE192 pKa = 4.12TNRR195 pKa = 11.84LQSSQAAGNEE205 pKa = 4.19SEE207 pKa = 4.77VPQNEE212 pKa = 4.14MPIRR216 pKa = 11.84NPNNRR221 pKa = 11.84GRR223 pKa = 11.84GRR225 pKa = 11.84SNRR228 pKa = 11.84RR229 pKa = 11.84RR230 pKa = 11.84GRR232 pKa = 11.84QRR234 pKa = 11.84PRR236 pKa = 11.84STTALPRR243 pKa = 11.84QTSQTRR249 pKa = 11.84GRR251 pKa = 11.84IRR253 pKa = 11.84QGSPNSAPNIDD264 pKa = 3.73PATTVGRR271 pKa = 11.84VRR273 pKa = 11.84NEE275 pKa = 3.68VQTRR279 pKa = 11.84RR280 pKa = 11.84ATAPNRR286 pKa = 11.84RR287 pKa = 11.84GSKK290 pKa = 9.12GARR293 pKa = 11.84HH294 pKa = 6.05FRR296 pKa = 11.84GQLL299 pKa = 3.12
MM1 pKa = 7.31EE2 pKa = 5.28AATLVVADD10 pKa = 4.08NQMDD14 pKa = 3.92RR15 pKa = 11.84NKK17 pKa = 9.95TYY19 pKa = 11.21ARR21 pKa = 11.84VVQILRR27 pKa = 11.84SRR29 pKa = 11.84MASRR33 pKa = 11.84QADD36 pKa = 3.52MTILSEE42 pKa = 4.19VATITLRR49 pKa = 11.84LADD52 pKa = 4.76NINRR56 pKa = 11.84DD57 pKa = 3.54AVLSGNFRR65 pKa = 11.84GTILDD70 pKa = 3.67YY71 pKa = 10.53FDD73 pKa = 3.39RR74 pKa = 11.84RR75 pKa = 11.84GWVAYY80 pKa = 9.15YY81 pKa = 10.35LIAPLGTWLPRR92 pKa = 11.84SVWGWMLSKK101 pKa = 8.33MTSRR105 pKa = 11.84EE106 pKa = 3.62HH107 pKa = 7.19RR108 pKa = 11.84FNRR111 pKa = 11.84WVWPHH116 pKa = 5.83VSATHH121 pKa = 6.92CSRR124 pKa = 11.84IMSADD129 pKa = 3.06EE130 pKa = 3.79LQRR133 pKa = 11.84IRR135 pKa = 11.84GALTAIDD142 pKa = 3.93PGARR146 pKa = 11.84QPPFQEE152 pKa = 4.49PGLVLNAIHH161 pKa = 7.11SDD163 pKa = 3.35PGTDD167 pKa = 3.17SDD169 pKa = 4.27GTHH172 pKa = 5.57GRR174 pKa = 11.84PRR176 pKa = 11.84EE177 pKa = 3.83RR178 pKa = 11.84DD179 pKa = 3.56RR180 pKa = 11.84IPSNPSSSTRR190 pKa = 11.84AEE192 pKa = 4.12TNRR195 pKa = 11.84LQSSQAAGNEE205 pKa = 4.19SEE207 pKa = 4.77VPQNEE212 pKa = 4.14MPIRR216 pKa = 11.84NPNNRR221 pKa = 11.84GRR223 pKa = 11.84GRR225 pKa = 11.84SNRR228 pKa = 11.84RR229 pKa = 11.84RR230 pKa = 11.84GRR232 pKa = 11.84QRR234 pKa = 11.84PRR236 pKa = 11.84STTALPRR243 pKa = 11.84QTSQTRR249 pKa = 11.84GRR251 pKa = 11.84IRR253 pKa = 11.84QGSPNSAPNIDD264 pKa = 3.73PATTVGRR271 pKa = 11.84VRR273 pKa = 11.84NEE275 pKa = 3.68VQTRR279 pKa = 11.84RR280 pKa = 11.84ATAPNRR286 pKa = 11.84RR287 pKa = 11.84GSKK290 pKa = 9.12GARR293 pKa = 11.84HH294 pKa = 6.05FRR296 pKa = 11.84GQLL299 pKa = 3.12
Molecular weight: 33.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1216 |
299 |
557 |
405.3 |
45.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.059 ± 0.601 | 1.48 ± 0.608 |
4.688 ± 0.21 | 4.359 ± 1.009 |
3.618 ± 0.805 | 6.003 ± 0.759 |
1.727 ± 0.428 | 5.345 ± 0.343 |
3.947 ± 0.986 | 8.553 ± 0.908 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.207 ± 0.239 | 4.03 ± 0.979 |
6.497 ± 0.239 | 4.77 ± 0.263 |
7.813 ± 2.596 | 7.73 ± 1.709 |
6.086 ± 0.555 | 7.072 ± 0.724 |
1.809 ± 0.147 | 3.207 ± 0.65 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
