
Defluviimonas aquaemixtae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Defluviimonas
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
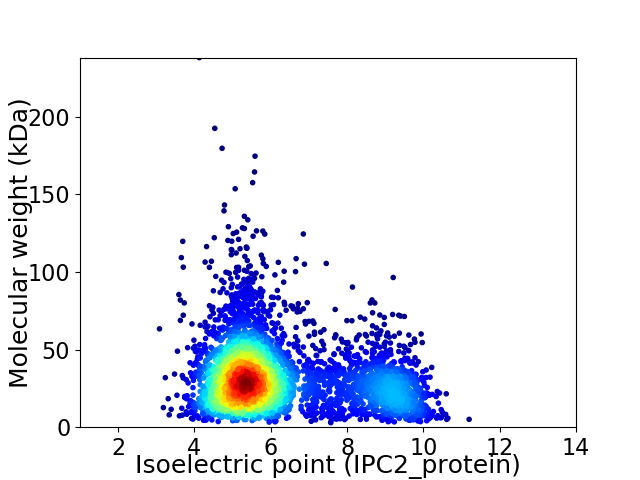
Virtual 2D-PAGE plot for 4129 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
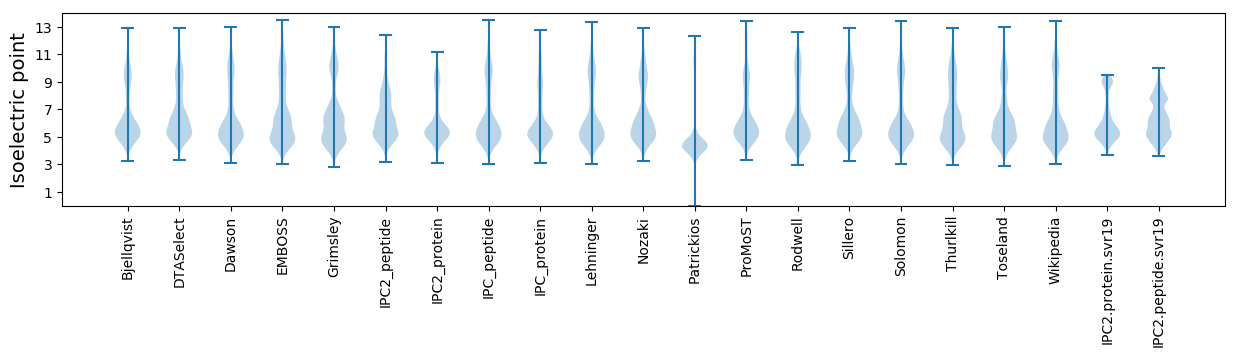
Protein with the lowest isoelectric point:
>tr|A0A2R8BM21|A0A2R8BM21_9RHOB PNPLA domain-containing protein OS=Defluviimonas aquaemixtae OX=1542388 GN=DEA8626_03508 PE=4 SV=1
MM1 pKa = 6.74STAIKK6 pKa = 10.35FDD8 pKa = 4.04FSQAAGEE15 pKa = 4.36EE16 pKa = 4.49IEE18 pKa = 4.79SSLFGANMIFTKK30 pKa = 10.83NILDD34 pKa = 4.06NNSSPTLDD42 pKa = 3.6NLNVSNIRR50 pKa = 11.84YY51 pKa = 8.6PGGSVTEE58 pKa = 4.25FNFDD62 pKa = 3.45YY63 pKa = 10.42RR64 pKa = 11.84NPDD67 pKa = 2.66IAVNDD72 pKa = 3.68NSLHH76 pKa = 6.13YY77 pKa = 11.26DD78 pKa = 3.42NDD80 pKa = 3.21QDD82 pKa = 4.12TAPTPLLPGEE92 pKa = 4.48FVPLTSFLDD101 pKa = 3.73WVEE104 pKa = 4.25STGKK108 pKa = 10.04SATVVLPTRR117 pKa = 11.84NLVDD121 pKa = 3.52QSTQQGTLPRR131 pKa = 11.84AINIEE136 pKa = 4.26AIEE139 pKa = 4.16DD140 pKa = 3.71LKK142 pKa = 11.45KK143 pKa = 10.78FLLDD147 pKa = 3.85LLSDD151 pKa = 3.54VDD153 pKa = 4.53SNGNAMQNADD163 pKa = 3.43IAALEE168 pKa = 4.22IGNEE172 pKa = 4.09YY173 pKa = 9.91WSLGSMTAAEE183 pKa = 4.22YY184 pKa = 10.62GAVVDD189 pKa = 4.76AVIAVIEE196 pKa = 4.53GVFEE200 pKa = 3.96EE201 pKa = 5.0LGISSQEE208 pKa = 3.97APDD211 pKa = 3.85VLVQMGSPWGVEE223 pKa = 4.24FQSGGAYY230 pKa = 10.3YY231 pKa = 10.6GITADD236 pKa = 3.55SPQYY240 pKa = 10.97ILDD243 pKa = 3.67QYY245 pKa = 11.62GLTDD249 pKa = 4.27SDD251 pKa = 4.07FLNDD255 pKa = 5.59GSLKK259 pKa = 9.7WLSKK263 pKa = 10.64INLVNLDD270 pKa = 4.13IIRR273 pKa = 11.84GISEE277 pKa = 4.13DD278 pKa = 3.29SRR280 pKa = 11.84NSVDD284 pKa = 3.8GVVDD288 pKa = 3.41HH289 pKa = 6.7YY290 pKa = 11.57YY291 pKa = 11.04YY292 pKa = 11.06HH293 pKa = 6.12NTDD296 pKa = 3.0NALSYY301 pKa = 10.88TSGSINYY308 pKa = 8.7IDD310 pKa = 4.2RR311 pKa = 11.84DD312 pKa = 3.77YY313 pKa = 11.46AIWSSNFGRR322 pKa = 11.84EE323 pKa = 3.88LDD325 pKa = 3.67LHH327 pKa = 4.91ITEE330 pKa = 4.92WNVKK334 pKa = 7.08TANLNQLGLKK344 pKa = 9.1GAGVILEE351 pKa = 4.09QVEE354 pKa = 4.14NLVRR358 pKa = 11.84LGVDD362 pKa = 3.45SAFVWPYY369 pKa = 9.29QHH371 pKa = 6.24NTRR374 pKa = 11.84NDD376 pKa = 3.59LAGSPGDD383 pKa = 3.98DD384 pKa = 3.36TSLTPLGAAFKK395 pKa = 10.31LAAEE399 pKa = 4.34SLVGTRR405 pKa = 11.84RR406 pKa = 11.84LDD408 pKa = 3.6SNISGGEE415 pKa = 4.04LEE417 pKa = 4.43VNAYY421 pKa = 9.12SASEE425 pKa = 3.98KK426 pKa = 10.58FVFFVSSRR434 pKa = 11.84SDD436 pKa = 3.23TQQSIEE442 pKa = 4.17IDD444 pKa = 3.57FGDD447 pKa = 3.88LVSSYY452 pKa = 11.14VSVSGVKK459 pKa = 9.87IGVDD463 pKa = 3.94LSTSDD468 pKa = 3.24GRR470 pKa = 11.84HH471 pKa = 4.48WTPSGISQVEE481 pKa = 4.12YY482 pKa = 10.58FNEE485 pKa = 4.05SDD487 pKa = 3.66VQAQLSQYY495 pKa = 11.02SEE497 pKa = 5.05DD498 pKa = 4.1EE499 pKa = 4.48LGTAANVSFVLDD511 pKa = 4.02PYY513 pKa = 11.3EE514 pKa = 4.34VMRR517 pKa = 11.84LEE519 pKa = 4.25FVLDD523 pKa = 3.75AARR526 pKa = 11.84QFSGASGNDD535 pKa = 3.11TWSGGAGYY543 pKa = 10.36DD544 pKa = 3.46VAFGSSGDD552 pKa = 3.75DD553 pKa = 3.17TLLGFDD559 pKa = 5.23GNDD562 pKa = 3.21TLAGDD567 pKa = 4.65DD568 pKa = 4.8GRR570 pKa = 11.84DD571 pKa = 3.4HH572 pKa = 6.94LAGGGGHH579 pKa = 7.36DD580 pKa = 4.58EE581 pKa = 4.49LNGGAGDD588 pKa = 3.78DD589 pKa = 4.56WITGDD594 pKa = 4.41SGNDD598 pKa = 3.67TIEE601 pKa = 4.41GGAGADD607 pKa = 3.62TLIGGSGIDD616 pKa = 3.46TVNYY620 pKa = 8.74YY621 pKa = 10.38YY622 pKa = 11.07SSGGVTIDD630 pKa = 3.89LLTNAVSGGWAADD643 pKa = 3.48DD644 pKa = 4.35TISGFEE650 pKa = 3.66RR651 pKa = 11.84AYY653 pKa = 10.73GSNSGNDD660 pKa = 3.3VLKK663 pKa = 10.75GSNGANVLRR672 pKa = 11.84GYY674 pKa = 10.2GGNDD678 pKa = 2.5IVYY681 pKa = 10.5DD682 pKa = 4.0RR683 pKa = 11.84GGDD686 pKa = 3.74DD687 pKa = 4.68YY688 pKa = 12.23VDD690 pKa = 4.52LGDD693 pKa = 4.78GNDD696 pKa = 3.54AVIVGNGADD705 pKa = 3.73TLIGGSGIDD714 pKa = 3.46TVNYY718 pKa = 8.74YY719 pKa = 10.38YY720 pKa = 11.07SSGGVTIDD728 pKa = 3.89LLTNAVSGGWAADD741 pKa = 3.48DD742 pKa = 4.35TISGFEE748 pKa = 3.66RR749 pKa = 11.84AYY751 pKa = 10.73GSNSGNDD758 pKa = 3.3VLKK761 pKa = 10.75GSNGANVLRR770 pKa = 11.84GYY772 pKa = 10.2GGNDD776 pKa = 2.5IVYY779 pKa = 10.5DD780 pKa = 4.0RR781 pKa = 11.84GGDD784 pKa = 3.74DD785 pKa = 4.68YY786 pKa = 12.23VDD788 pKa = 4.52LGDD791 pKa = 4.78GNDD794 pKa = 3.54AVIVGNGADD803 pKa = 3.73TLIGGSGIDD812 pKa = 3.46TVNYY816 pKa = 8.74YY817 pKa = 10.38YY818 pKa = 11.07SSGGVTIDD826 pKa = 3.89LLTNAVSGGWAADD839 pKa = 3.48DD840 pKa = 4.35TISGFEE846 pKa = 3.66RR847 pKa = 11.84AYY849 pKa = 10.73GSNSGNDD856 pKa = 3.3VLKK859 pKa = 10.75GSNGANVLRR868 pKa = 11.84GYY870 pKa = 10.3GGNDD874 pKa = 3.09RR875 pKa = 11.84LDD877 pKa = 3.37GRR879 pKa = 11.84AGNDD883 pKa = 3.41YY884 pKa = 11.13LDD886 pKa = 4.51GGSGGDD892 pKa = 3.47TLTGGVGADD901 pKa = 2.81HH902 pKa = 6.97FVFHH906 pKa = 6.55KK907 pKa = 9.83TYY909 pKa = 10.45GADD912 pKa = 4.19LITDD916 pKa = 4.25FAYY919 pKa = 10.45GLDD922 pKa = 3.89EE923 pKa = 5.02LVLDD927 pKa = 4.16HH928 pKa = 6.67SLWGGGLTEE937 pKa = 4.36AQVISIYY944 pKa = 10.95ASVIGEE950 pKa = 4.07NLVLDD955 pKa = 4.46FGGGDD960 pKa = 3.86TITLQGIASPGLLVDD975 pKa = 6.14DD976 pKa = 4.52ILIVV980 pKa = 3.44
MM1 pKa = 6.74STAIKK6 pKa = 10.35FDD8 pKa = 4.04FSQAAGEE15 pKa = 4.36EE16 pKa = 4.49IEE18 pKa = 4.79SSLFGANMIFTKK30 pKa = 10.83NILDD34 pKa = 4.06NNSSPTLDD42 pKa = 3.6NLNVSNIRR50 pKa = 11.84YY51 pKa = 8.6PGGSVTEE58 pKa = 4.25FNFDD62 pKa = 3.45YY63 pKa = 10.42RR64 pKa = 11.84NPDD67 pKa = 2.66IAVNDD72 pKa = 3.68NSLHH76 pKa = 6.13YY77 pKa = 11.26DD78 pKa = 3.42NDD80 pKa = 3.21QDD82 pKa = 4.12TAPTPLLPGEE92 pKa = 4.48FVPLTSFLDD101 pKa = 3.73WVEE104 pKa = 4.25STGKK108 pKa = 10.04SATVVLPTRR117 pKa = 11.84NLVDD121 pKa = 3.52QSTQQGTLPRR131 pKa = 11.84AINIEE136 pKa = 4.26AIEE139 pKa = 4.16DD140 pKa = 3.71LKK142 pKa = 11.45KK143 pKa = 10.78FLLDD147 pKa = 3.85LLSDD151 pKa = 3.54VDD153 pKa = 4.53SNGNAMQNADD163 pKa = 3.43IAALEE168 pKa = 4.22IGNEE172 pKa = 4.09YY173 pKa = 9.91WSLGSMTAAEE183 pKa = 4.22YY184 pKa = 10.62GAVVDD189 pKa = 4.76AVIAVIEE196 pKa = 4.53GVFEE200 pKa = 3.96EE201 pKa = 5.0LGISSQEE208 pKa = 3.97APDD211 pKa = 3.85VLVQMGSPWGVEE223 pKa = 4.24FQSGGAYY230 pKa = 10.3YY231 pKa = 10.6GITADD236 pKa = 3.55SPQYY240 pKa = 10.97ILDD243 pKa = 3.67QYY245 pKa = 11.62GLTDD249 pKa = 4.27SDD251 pKa = 4.07FLNDD255 pKa = 5.59GSLKK259 pKa = 9.7WLSKK263 pKa = 10.64INLVNLDD270 pKa = 4.13IIRR273 pKa = 11.84GISEE277 pKa = 4.13DD278 pKa = 3.29SRR280 pKa = 11.84NSVDD284 pKa = 3.8GVVDD288 pKa = 3.41HH289 pKa = 6.7YY290 pKa = 11.57YY291 pKa = 11.04YY292 pKa = 11.06HH293 pKa = 6.12NTDD296 pKa = 3.0NALSYY301 pKa = 10.88TSGSINYY308 pKa = 8.7IDD310 pKa = 4.2RR311 pKa = 11.84DD312 pKa = 3.77YY313 pKa = 11.46AIWSSNFGRR322 pKa = 11.84EE323 pKa = 3.88LDD325 pKa = 3.67LHH327 pKa = 4.91ITEE330 pKa = 4.92WNVKK334 pKa = 7.08TANLNQLGLKK344 pKa = 9.1GAGVILEE351 pKa = 4.09QVEE354 pKa = 4.14NLVRR358 pKa = 11.84LGVDD362 pKa = 3.45SAFVWPYY369 pKa = 9.29QHH371 pKa = 6.24NTRR374 pKa = 11.84NDD376 pKa = 3.59LAGSPGDD383 pKa = 3.98DD384 pKa = 3.36TSLTPLGAAFKK395 pKa = 10.31LAAEE399 pKa = 4.34SLVGTRR405 pKa = 11.84RR406 pKa = 11.84LDD408 pKa = 3.6SNISGGEE415 pKa = 4.04LEE417 pKa = 4.43VNAYY421 pKa = 9.12SASEE425 pKa = 3.98KK426 pKa = 10.58FVFFVSSRR434 pKa = 11.84SDD436 pKa = 3.23TQQSIEE442 pKa = 4.17IDD444 pKa = 3.57FGDD447 pKa = 3.88LVSSYY452 pKa = 11.14VSVSGVKK459 pKa = 9.87IGVDD463 pKa = 3.94LSTSDD468 pKa = 3.24GRR470 pKa = 11.84HH471 pKa = 4.48WTPSGISQVEE481 pKa = 4.12YY482 pKa = 10.58FNEE485 pKa = 4.05SDD487 pKa = 3.66VQAQLSQYY495 pKa = 11.02SEE497 pKa = 5.05DD498 pKa = 4.1EE499 pKa = 4.48LGTAANVSFVLDD511 pKa = 4.02PYY513 pKa = 11.3EE514 pKa = 4.34VMRR517 pKa = 11.84LEE519 pKa = 4.25FVLDD523 pKa = 3.75AARR526 pKa = 11.84QFSGASGNDD535 pKa = 3.11TWSGGAGYY543 pKa = 10.36DD544 pKa = 3.46VAFGSSGDD552 pKa = 3.75DD553 pKa = 3.17TLLGFDD559 pKa = 5.23GNDD562 pKa = 3.21TLAGDD567 pKa = 4.65DD568 pKa = 4.8GRR570 pKa = 11.84DD571 pKa = 3.4HH572 pKa = 6.94LAGGGGHH579 pKa = 7.36DD580 pKa = 4.58EE581 pKa = 4.49LNGGAGDD588 pKa = 3.78DD589 pKa = 4.56WITGDD594 pKa = 4.41SGNDD598 pKa = 3.67TIEE601 pKa = 4.41GGAGADD607 pKa = 3.62TLIGGSGIDD616 pKa = 3.46TVNYY620 pKa = 8.74YY621 pKa = 10.38YY622 pKa = 11.07SSGGVTIDD630 pKa = 3.89LLTNAVSGGWAADD643 pKa = 3.48DD644 pKa = 4.35TISGFEE650 pKa = 3.66RR651 pKa = 11.84AYY653 pKa = 10.73GSNSGNDD660 pKa = 3.3VLKK663 pKa = 10.75GSNGANVLRR672 pKa = 11.84GYY674 pKa = 10.2GGNDD678 pKa = 2.5IVYY681 pKa = 10.5DD682 pKa = 4.0RR683 pKa = 11.84GGDD686 pKa = 3.74DD687 pKa = 4.68YY688 pKa = 12.23VDD690 pKa = 4.52LGDD693 pKa = 4.78GNDD696 pKa = 3.54AVIVGNGADD705 pKa = 3.73TLIGGSGIDD714 pKa = 3.46TVNYY718 pKa = 8.74YY719 pKa = 10.38YY720 pKa = 11.07SSGGVTIDD728 pKa = 3.89LLTNAVSGGWAADD741 pKa = 3.48DD742 pKa = 4.35TISGFEE748 pKa = 3.66RR749 pKa = 11.84AYY751 pKa = 10.73GSNSGNDD758 pKa = 3.3VLKK761 pKa = 10.75GSNGANVLRR770 pKa = 11.84GYY772 pKa = 10.2GGNDD776 pKa = 2.5IVYY779 pKa = 10.5DD780 pKa = 4.0RR781 pKa = 11.84GGDD784 pKa = 3.74DD785 pKa = 4.68YY786 pKa = 12.23VDD788 pKa = 4.52LGDD791 pKa = 4.78GNDD794 pKa = 3.54AVIVGNGADD803 pKa = 3.73TLIGGSGIDD812 pKa = 3.46TVNYY816 pKa = 8.74YY817 pKa = 10.38YY818 pKa = 11.07SSGGVTIDD826 pKa = 3.89LLTNAVSGGWAADD839 pKa = 3.48DD840 pKa = 4.35TISGFEE846 pKa = 3.66RR847 pKa = 11.84AYY849 pKa = 10.73GSNSGNDD856 pKa = 3.3VLKK859 pKa = 10.75GSNGANVLRR868 pKa = 11.84GYY870 pKa = 10.3GGNDD874 pKa = 3.09RR875 pKa = 11.84LDD877 pKa = 3.37GRR879 pKa = 11.84AGNDD883 pKa = 3.41YY884 pKa = 11.13LDD886 pKa = 4.51GGSGGDD892 pKa = 3.47TLTGGVGADD901 pKa = 2.81HH902 pKa = 6.97FVFHH906 pKa = 6.55KK907 pKa = 9.83TYY909 pKa = 10.45GADD912 pKa = 4.19LITDD916 pKa = 4.25FAYY919 pKa = 10.45GLDD922 pKa = 3.89EE923 pKa = 5.02LVLDD927 pKa = 4.16HH928 pKa = 6.67SLWGGGLTEE937 pKa = 4.36AQVISIYY944 pKa = 10.95ASVIGEE950 pKa = 4.07NLVLDD955 pKa = 4.46FGGGDD960 pKa = 3.86TITLQGIASPGLLVDD975 pKa = 6.14DD976 pKa = 4.52ILIVV980 pKa = 3.44
Molecular weight: 103.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R8B1Q6|A0A2R8B1Q6_9RHOB Uncharacterized protein OS=Defluviimonas aquaemixtae OX=1542388 GN=DEA8626_00081 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.56GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.56GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1262290 |
30 |
2386 |
305.7 |
33.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.849 ± 0.055 | 0.906 ± 0.013 |
5.924 ± 0.031 | 6.182 ± 0.042 |
3.716 ± 0.024 | 8.908 ± 0.038 |
2.062 ± 0.02 | 5.081 ± 0.029 |
3.038 ± 0.036 | 9.909 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.627 ± 0.018 | 2.412 ± 0.023 |
5.218 ± 0.027 | 2.678 ± 0.023 |
7.269 ± 0.041 | 4.999 ± 0.025 |
5.259 ± 0.029 | 7.327 ± 0.03 |
1.449 ± 0.016 | 2.188 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
