
Wuchereria bancrofti
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Nematoda; Chromadorea; Rhabditida; Spirurina; Spiruromorpha; Filarioidea; Onchocercidae; Wuchereria
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
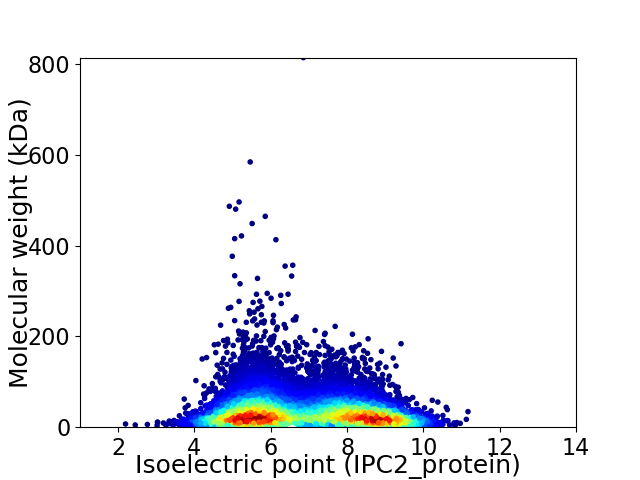
Virtual 2D-PAGE plot for 13000 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
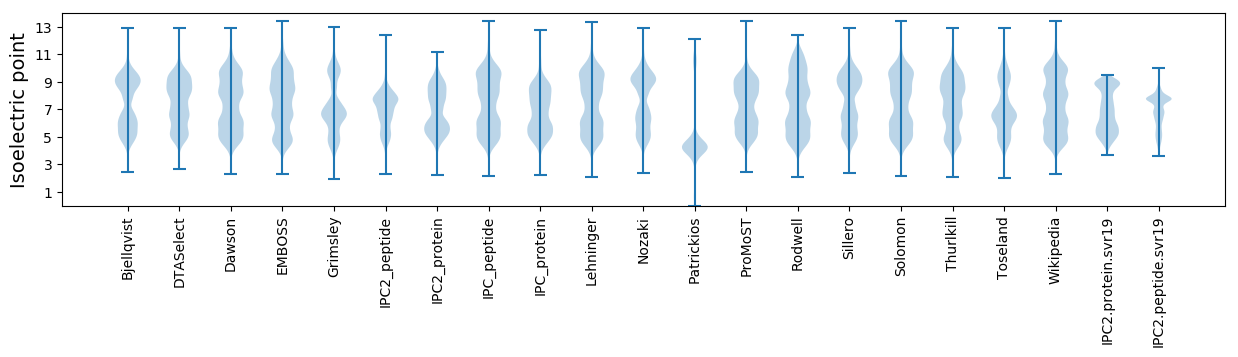
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3P7DX92|A0A3P7DX92_WUCBA RBR-type E3 ubiquitin transferase OS=Wuchereria bancrofti OX=6293 GN=WBA_LOCUS1435 PE=4 SV=1
MM1 pKa = 7.7NSLQIKK7 pKa = 9.42WSLNYY12 pKa = 10.25NAITSLIKK20 pKa = 10.5FKK22 pKa = 11.04YY23 pKa = 9.92LAFISLLYY31 pKa = 10.47RR32 pKa = 11.84FTFSSVNNDD41 pKa = 3.01VNNGGGGDD49 pKa = 5.02DD50 pKa = 6.13DD51 pKa = 6.84DD52 pKa = 7.23DD53 pKa = 7.21DD54 pKa = 6.73YY55 pKa = 12.16DD56 pKa = 6.59DD57 pKa = 6.57GDD59 pKa = 6.17DD60 pKa = 6.41DD61 pKa = 6.96DD62 pKa = 7.63DD63 pKa = 6.74DD64 pKa = 7.15DD65 pKa = 5.94NDD67 pKa = 4.68EE68 pKa = 5.45DD69 pKa = 4.97NDD71 pKa = 5.33DD72 pKa = 3.7INIMYY77 pKa = 9.97FLEE80 pKa = 4.29STLPVHH86 pKa = 6.22YY87 pKa = 9.67FYY89 pKa = 11.41LSTDD93 pKa = 2.94LRR95 pKa = 11.84FVHH98 pKa = 6.86IFSQITSIHH107 pKa = 6.0VKK109 pKa = 9.89TII111 pKa = 3.16
MM1 pKa = 7.7NSLQIKK7 pKa = 9.42WSLNYY12 pKa = 10.25NAITSLIKK20 pKa = 10.5FKK22 pKa = 11.04YY23 pKa = 9.92LAFISLLYY31 pKa = 10.47RR32 pKa = 11.84FTFSSVNNDD41 pKa = 3.01VNNGGGGDD49 pKa = 5.02DD50 pKa = 6.13DD51 pKa = 6.84DD52 pKa = 7.23DD53 pKa = 7.21DD54 pKa = 6.73YY55 pKa = 12.16DD56 pKa = 6.59DD57 pKa = 6.57GDD59 pKa = 6.17DD60 pKa = 6.41DD61 pKa = 6.96DD62 pKa = 7.63DD63 pKa = 6.74DD64 pKa = 7.15DD65 pKa = 5.94NDD67 pKa = 4.68EE68 pKa = 5.45DD69 pKa = 4.97NDD71 pKa = 5.33DD72 pKa = 3.7INIMYY77 pKa = 9.97FLEE80 pKa = 4.29STLPVHH86 pKa = 6.22YY87 pKa = 9.67FYY89 pKa = 11.41LSTDD93 pKa = 2.94LRR95 pKa = 11.84FVHH98 pKa = 6.86IFSQITSIHH107 pKa = 6.0VKK109 pKa = 9.89TII111 pKa = 3.16
Molecular weight: 12.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3P7E8P8|A0A3P7E8P8_WUCBA Uncharacterized protein OS=Wuchereria bancrofti OX=6293 GN=WBA_LOCUS5705 PE=3 SV=1
MM1 pKa = 6.71VRR3 pKa = 11.84AVRR6 pKa = 11.84ALRR9 pKa = 11.84RR10 pKa = 11.84PLRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84TARR18 pKa = 11.84VHH20 pKa = 6.25LAPGGLLRR28 pKa = 11.84RR29 pKa = 11.84IGAGALGWAVPLRR42 pKa = 11.84PRR44 pKa = 11.84LPRR47 pKa = 11.84RR48 pKa = 11.84LAPVRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84ARR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84ARR61 pKa = 11.84DD62 pKa = 3.33DD63 pKa = 3.43GAPRR67 pKa = 11.84DD68 pKa = 4.05RR69 pKa = 11.84ARR71 pKa = 11.84HH72 pKa = 4.93RR73 pKa = 11.84APRR76 pKa = 11.84AARR79 pKa = 11.84PGAGPGARR87 pKa = 11.84GRR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84RR92 pKa = 11.84ALRR95 pKa = 11.84DD96 pKa = 3.18PRR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84EE101 pKa = 3.74AAGDD105 pKa = 3.62APHH108 pKa = 7.57PLRR111 pKa = 11.84LPPRR115 pKa = 11.84PPAGRR120 pKa = 11.84VGQRR124 pKa = 11.84QRR126 pKa = 11.84PGAGRR131 pKa = 11.84GAQPRR136 pKa = 11.84RR137 pKa = 11.84QGRR140 pKa = 11.84GGPGRR145 pKa = 11.84RR146 pKa = 11.84VRR148 pKa = 11.84RR149 pKa = 11.84PRR151 pKa = 11.84LARR154 pKa = 11.84RR155 pKa = 11.84RR156 pKa = 11.84PTSGARR162 pKa = 11.84RR163 pKa = 11.84PRR165 pKa = 11.84DD166 pKa = 3.5GAGGPHH172 pKa = 6.95RR173 pKa = 11.84AGAAARR179 pKa = 11.84AAGAHH184 pKa = 5.04HH185 pKa = 7.44RR186 pKa = 11.84GRR188 pKa = 11.84ARR190 pKa = 11.84RR191 pKa = 11.84GPPARR196 pKa = 11.84GPGRR200 pKa = 11.84HH201 pKa = 4.94QPGPVRR207 pKa = 11.84ARR209 pKa = 11.84RR210 pKa = 11.84PRR212 pKa = 11.84HH213 pKa = 4.47QHH215 pKa = 5.9PALAPLPAALDD226 pKa = 3.43GDD228 pKa = 3.62RR229 pKa = 11.84RR230 pKa = 11.84LARR233 pKa = 11.84RR234 pKa = 11.84GAGAPRR240 pKa = 11.84PRR242 pKa = 11.84RR243 pKa = 11.84GLGAARR249 pKa = 11.84RR250 pKa = 11.84RR251 pKa = 11.84LAAHH255 pKa = 6.89LRR257 pKa = 11.84PGHH260 pKa = 6.13RR261 pKa = 11.84GRR263 pKa = 11.84RR264 pKa = 11.84GLVAGGHH271 pKa = 4.84QGRR274 pKa = 11.84RR275 pKa = 11.84TARR278 pKa = 11.84ARR280 pKa = 11.84RR281 pKa = 11.84RR282 pKa = 11.84RR283 pKa = 11.84RR284 pKa = 11.84GGGGARR290 pKa = 11.84GRR292 pKa = 11.84TPRR295 pKa = 11.84LGGRR299 pKa = 11.84RR300 pKa = 11.84RR301 pKa = 11.84PAAGGGARR309 pKa = 11.84AGRR312 pKa = 11.84AVGGTT317 pKa = 3.52
MM1 pKa = 6.71VRR3 pKa = 11.84AVRR6 pKa = 11.84ALRR9 pKa = 11.84RR10 pKa = 11.84PLRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84TARR18 pKa = 11.84VHH20 pKa = 6.25LAPGGLLRR28 pKa = 11.84RR29 pKa = 11.84IGAGALGWAVPLRR42 pKa = 11.84PRR44 pKa = 11.84LPRR47 pKa = 11.84RR48 pKa = 11.84LAPVRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84ARR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84ARR61 pKa = 11.84DD62 pKa = 3.33DD63 pKa = 3.43GAPRR67 pKa = 11.84DD68 pKa = 4.05RR69 pKa = 11.84ARR71 pKa = 11.84HH72 pKa = 4.93RR73 pKa = 11.84APRR76 pKa = 11.84AARR79 pKa = 11.84PGAGPGARR87 pKa = 11.84GRR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84RR92 pKa = 11.84ALRR95 pKa = 11.84DD96 pKa = 3.18PRR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84EE101 pKa = 3.74AAGDD105 pKa = 3.62APHH108 pKa = 7.57PLRR111 pKa = 11.84LPPRR115 pKa = 11.84PPAGRR120 pKa = 11.84VGQRR124 pKa = 11.84QRR126 pKa = 11.84PGAGRR131 pKa = 11.84GAQPRR136 pKa = 11.84RR137 pKa = 11.84QGRR140 pKa = 11.84GGPGRR145 pKa = 11.84RR146 pKa = 11.84VRR148 pKa = 11.84RR149 pKa = 11.84PRR151 pKa = 11.84LARR154 pKa = 11.84RR155 pKa = 11.84RR156 pKa = 11.84PTSGARR162 pKa = 11.84RR163 pKa = 11.84PRR165 pKa = 11.84DD166 pKa = 3.5GAGGPHH172 pKa = 6.95RR173 pKa = 11.84AGAAARR179 pKa = 11.84AAGAHH184 pKa = 5.04HH185 pKa = 7.44RR186 pKa = 11.84GRR188 pKa = 11.84ARR190 pKa = 11.84RR191 pKa = 11.84GPPARR196 pKa = 11.84GPGRR200 pKa = 11.84HH201 pKa = 4.94QPGPVRR207 pKa = 11.84ARR209 pKa = 11.84RR210 pKa = 11.84PRR212 pKa = 11.84HH213 pKa = 4.47QHH215 pKa = 5.9PALAPLPAALDD226 pKa = 3.43GDD228 pKa = 3.62RR229 pKa = 11.84RR230 pKa = 11.84LARR233 pKa = 11.84RR234 pKa = 11.84GAGAPRR240 pKa = 11.84PRR242 pKa = 11.84RR243 pKa = 11.84GLGAARR249 pKa = 11.84RR250 pKa = 11.84RR251 pKa = 11.84LAAHH255 pKa = 6.89LRR257 pKa = 11.84PGHH260 pKa = 6.13RR261 pKa = 11.84GRR263 pKa = 11.84RR264 pKa = 11.84GLVAGGHH271 pKa = 4.84QGRR274 pKa = 11.84RR275 pKa = 11.84TARR278 pKa = 11.84ARR280 pKa = 11.84RR281 pKa = 11.84RR282 pKa = 11.84RR283 pKa = 11.84RR284 pKa = 11.84GGGGARR290 pKa = 11.84GRR292 pKa = 11.84TPRR295 pKa = 11.84LGGRR299 pKa = 11.84RR300 pKa = 11.84RR301 pKa = 11.84PAAGGGARR309 pKa = 11.84AGRR312 pKa = 11.84AVGGTT317 pKa = 3.52
Molecular weight: 34.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4298790 |
29 |
7087 |
330.7 |
37.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.031 ± 0.022 | 2.218 ± 0.017 |
5.394 ± 0.016 | 6.627 ± 0.026 |
4.342 ± 0.018 | 4.914 ± 0.022 |
2.391 ± 0.009 | 6.693 ± 0.023 |
6.307 ± 0.023 | 9.404 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.544 ± 0.009 | 5.231 ± 0.017 |
4.061 ± 0.02 | 4.144 ± 0.018 |
5.687 ± 0.021 | 8.133 ± 0.025 |
5.587 ± 0.016 | 5.942 ± 0.019 |
1.1 ± 0.006 | 3.218 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
