
Macaca mulatta papillomavirus 7
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; unclassified Gammapapillomavirus
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
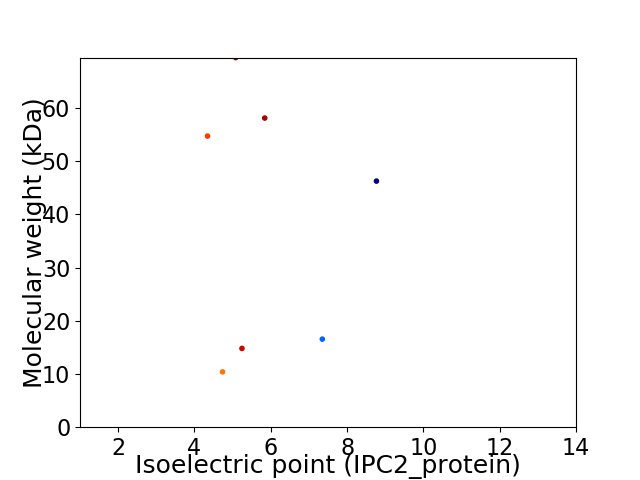
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A386JT45|A0A386JT45_9PAPI Regulatory protein E2 OS=Macaca mulatta papillomavirus 7 OX=2364644 GN=E2 PE=3 SV=1
MM1 pKa = 6.98YY2 pKa = 10.35KK3 pKa = 10.05VRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.57RR8 pKa = 11.84AAPEE12 pKa = 3.58DD13 pKa = 4.05LYY15 pKa = 10.45RR16 pKa = 11.84TCATGDD22 pKa = 3.71CPPDD26 pKa = 3.37VKK28 pKa = 11.19NKK30 pKa = 10.17IEE32 pKa = 4.26GNTLADD38 pKa = 4.69RR39 pKa = 11.84LLKK42 pKa = 9.79WFSSIIYY49 pKa = 10.16LGGLGIGTGRR59 pKa = 11.84GSGGSLGYY67 pKa = 10.32RR68 pKa = 11.84PIGGSGTRR76 pKa = 11.84PAPDD80 pKa = 4.38TIPIRR85 pKa = 11.84PAVPVDD91 pKa = 3.52PLGPPDD97 pKa = 3.78IVSVDD102 pKa = 3.92TIDD105 pKa = 3.59PAGPSIISEE114 pKa = 4.44TTTTTVVDD122 pKa = 3.83PSVIDD127 pKa = 3.65VASGGTGEE135 pKa = 4.79IEE137 pKa = 4.62HH138 pKa = 6.74GPIDD142 pKa = 4.98PISDD146 pKa = 2.95IGGTGGHH153 pKa = 5.84PTIITTDD160 pKa = 3.29NDD162 pKa = 3.67TVAVLDD168 pKa = 4.05VQPIPPPNRR177 pKa = 11.84AAIDD181 pKa = 3.73VSVGPGDD188 pKa = 3.79SSHH191 pKa = 7.34VSIITAVSHH200 pKa = 6.73PSPDD204 pKa = 2.74INVFVDD210 pKa = 3.97PSFAGEE216 pKa = 4.05TVGYY220 pKa = 10.52DD221 pKa = 3.94EE222 pKa = 6.54IPLQDD227 pKa = 4.06LGGLSEE233 pKa = 4.31FEE235 pKa = 3.98IDD237 pKa = 3.86EE238 pKa = 4.86GPTTSTPIQSLQRR251 pKa = 11.84VVGRR255 pKa = 11.84ARR257 pKa = 11.84NLYY260 pKa = 9.98HH261 pKa = 7.44RR262 pKa = 11.84FVQQIPTRR270 pKa = 11.84DD271 pKa = 3.43PAFLGQVSRR280 pKa = 11.84LAQFEE285 pKa = 4.59YY286 pKa = 10.64EE287 pKa = 4.8NPAFDD292 pKa = 4.58PDD294 pKa = 3.35ITVEE298 pKa = 4.07FQRR301 pKa = 11.84DD302 pKa = 3.38LEE304 pKa = 4.73AIEE307 pKa = 4.45AAPVSEE313 pKa = 4.28FQDD316 pKa = 2.77IRR318 pKa = 11.84ILHH321 pKa = 6.34RR322 pKa = 11.84PLYY325 pKa = 10.13SLNPEE330 pKa = 3.66GAVRR334 pKa = 11.84VSRR337 pKa = 11.84LGQRR341 pKa = 11.84GTITTRR347 pKa = 11.84SGLQIGQPVHH357 pKa = 6.71FYY359 pKa = 10.88YY360 pKa = 10.57DD361 pKa = 3.29VSEE364 pKa = 4.16ISPVDD369 pKa = 4.07SIEE372 pKa = 3.95LSPRR376 pKa = 11.84GEE378 pKa = 4.11FSGTTTVVDD387 pKa = 4.46ALAEE391 pKa = 4.3STFIDD396 pKa = 4.17PSEE399 pKa = 3.94ALQVFEE405 pKa = 5.35EE406 pKa = 4.48EE407 pKa = 4.06EE408 pKa = 4.36LYY410 pKa = 11.06DD411 pKa = 4.74RR412 pKa = 11.84LNEE415 pKa = 4.62DD416 pKa = 3.42FTNSQLVLTATDD428 pKa = 3.68EE429 pKa = 4.53LGEE432 pKa = 4.29SLTIPSLPPGTSLKK446 pKa = 10.76VFVDD450 pKa = 4.83DD451 pKa = 4.04YY452 pKa = 11.47SSGIIVNYY460 pKa = 7.82PTDD463 pKa = 3.73TTSVPSSTIVPSIPSIGPAIYY484 pKa = 9.57IDD486 pKa = 4.17SFGSDD491 pKa = 3.9YY492 pKa = 11.06YY493 pKa = 10.91LHH495 pKa = 7.43PSLIPRR501 pKa = 11.84KK502 pKa = 8.97RR503 pKa = 11.84KK504 pKa = 10.13RR505 pKa = 11.84MDD507 pKa = 3.0MFF509 pKa = 5.79
MM1 pKa = 6.98YY2 pKa = 10.35KK3 pKa = 10.05VRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.57RR8 pKa = 11.84AAPEE12 pKa = 3.58DD13 pKa = 4.05LYY15 pKa = 10.45RR16 pKa = 11.84TCATGDD22 pKa = 3.71CPPDD26 pKa = 3.37VKK28 pKa = 11.19NKK30 pKa = 10.17IEE32 pKa = 4.26GNTLADD38 pKa = 4.69RR39 pKa = 11.84LLKK42 pKa = 9.79WFSSIIYY49 pKa = 10.16LGGLGIGTGRR59 pKa = 11.84GSGGSLGYY67 pKa = 10.32RR68 pKa = 11.84PIGGSGTRR76 pKa = 11.84PAPDD80 pKa = 4.38TIPIRR85 pKa = 11.84PAVPVDD91 pKa = 3.52PLGPPDD97 pKa = 3.78IVSVDD102 pKa = 3.92TIDD105 pKa = 3.59PAGPSIISEE114 pKa = 4.44TTTTTVVDD122 pKa = 3.83PSVIDD127 pKa = 3.65VASGGTGEE135 pKa = 4.79IEE137 pKa = 4.62HH138 pKa = 6.74GPIDD142 pKa = 4.98PISDD146 pKa = 2.95IGGTGGHH153 pKa = 5.84PTIITTDD160 pKa = 3.29NDD162 pKa = 3.67TVAVLDD168 pKa = 4.05VQPIPPPNRR177 pKa = 11.84AAIDD181 pKa = 3.73VSVGPGDD188 pKa = 3.79SSHH191 pKa = 7.34VSIITAVSHH200 pKa = 6.73PSPDD204 pKa = 2.74INVFVDD210 pKa = 3.97PSFAGEE216 pKa = 4.05TVGYY220 pKa = 10.52DD221 pKa = 3.94EE222 pKa = 6.54IPLQDD227 pKa = 4.06LGGLSEE233 pKa = 4.31FEE235 pKa = 3.98IDD237 pKa = 3.86EE238 pKa = 4.86GPTTSTPIQSLQRR251 pKa = 11.84VVGRR255 pKa = 11.84ARR257 pKa = 11.84NLYY260 pKa = 9.98HH261 pKa = 7.44RR262 pKa = 11.84FVQQIPTRR270 pKa = 11.84DD271 pKa = 3.43PAFLGQVSRR280 pKa = 11.84LAQFEE285 pKa = 4.59YY286 pKa = 10.64EE287 pKa = 4.8NPAFDD292 pKa = 4.58PDD294 pKa = 3.35ITVEE298 pKa = 4.07FQRR301 pKa = 11.84DD302 pKa = 3.38LEE304 pKa = 4.73AIEE307 pKa = 4.45AAPVSEE313 pKa = 4.28FQDD316 pKa = 2.77IRR318 pKa = 11.84ILHH321 pKa = 6.34RR322 pKa = 11.84PLYY325 pKa = 10.13SLNPEE330 pKa = 3.66GAVRR334 pKa = 11.84VSRR337 pKa = 11.84LGQRR341 pKa = 11.84GTITTRR347 pKa = 11.84SGLQIGQPVHH357 pKa = 6.71FYY359 pKa = 10.88YY360 pKa = 10.57DD361 pKa = 3.29VSEE364 pKa = 4.16ISPVDD369 pKa = 4.07SIEE372 pKa = 3.95LSPRR376 pKa = 11.84GEE378 pKa = 4.11FSGTTTVVDD387 pKa = 4.46ALAEE391 pKa = 4.3STFIDD396 pKa = 4.17PSEE399 pKa = 3.94ALQVFEE405 pKa = 5.35EE406 pKa = 4.48EE407 pKa = 4.06EE408 pKa = 4.36LYY410 pKa = 11.06DD411 pKa = 4.74RR412 pKa = 11.84LNEE415 pKa = 4.62DD416 pKa = 3.42FTNSQLVLTATDD428 pKa = 3.68EE429 pKa = 4.53LGEE432 pKa = 4.29SLTIPSLPPGTSLKK446 pKa = 10.76VFVDD450 pKa = 4.83DD451 pKa = 4.04YY452 pKa = 11.47SSGIIVNYY460 pKa = 7.82PTDD463 pKa = 3.73TTSVPSSTIVPSIPSIGPAIYY484 pKa = 9.57IDD486 pKa = 4.17SFGSDD491 pKa = 3.9YY492 pKa = 11.06YY493 pKa = 10.91LHH495 pKa = 7.43PSLIPRR501 pKa = 11.84KK502 pKa = 8.97RR503 pKa = 11.84KK504 pKa = 10.13RR505 pKa = 11.84MDD507 pKa = 3.0MFF509 pKa = 5.79
Molecular weight: 54.73 kDa
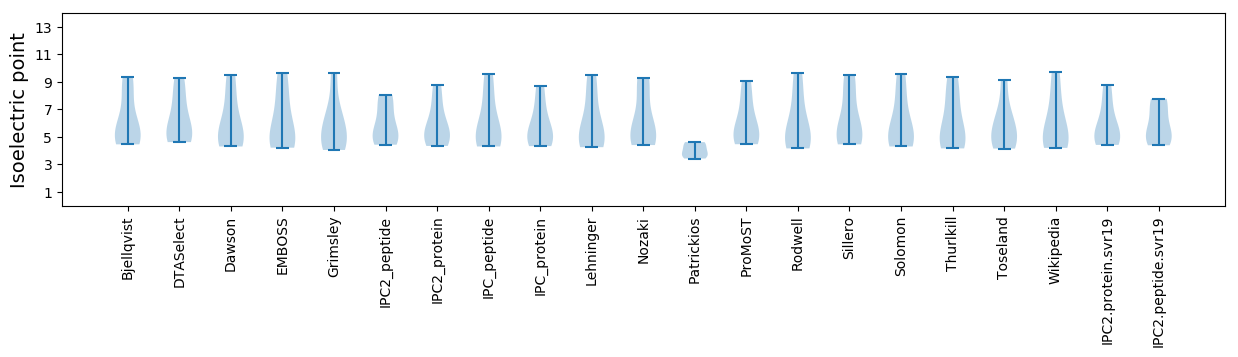
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A386JTE6|A0A386JTE6_9PAPI Protein E6 OS=Macaca mulatta papillomavirus 7 OX=2364644 GN=E6 PE=3 SV=1
MM1 pKa = 7.11EE2 pKa = 4.22TRR4 pKa = 11.84EE5 pKa = 4.04TLTARR10 pKa = 11.84FDD12 pKa = 3.73ALQEE16 pKa = 4.12QIMTLYY22 pKa = 10.52EE23 pKa = 4.0RR24 pKa = 11.84GAADD28 pKa = 3.94LQTQILHH35 pKa = 6.35WDD37 pKa = 4.09LVRR40 pKa = 11.84KK41 pKa = 9.92EE42 pKa = 4.11NVLLYY47 pKa = 10.06HH48 pKa = 6.09SRR50 pKa = 11.84KK51 pKa = 9.17QGFMSLGLQPTPALQVSEE69 pKa = 4.32YY70 pKa = 9.3RR71 pKa = 11.84AKK73 pKa = 10.59EE74 pKa = 4.11AIQMGILLNSLAKK87 pKa = 10.04SQYY90 pKa = 10.97ANEE93 pKa = 4.12RR94 pKa = 11.84WTLSDD99 pKa = 3.36TSAQLLLTEE108 pKa = 5.09PKK110 pKa = 10.2YY111 pKa = 10.75CFKK114 pKa = 10.83KK115 pKa = 10.19GAYY118 pKa = 8.05QVEE121 pKa = 4.98VYY123 pKa = 10.58FDD125 pKa = 3.55NDD127 pKa = 3.23EE128 pKa = 5.38ANMFPYY134 pKa = 10.43PNWNYY139 pKa = 10.2IYY141 pKa = 10.67YY142 pKa = 9.82QDD144 pKa = 5.05EE145 pKa = 4.12EE146 pKa = 4.51EE147 pKa = 4.37RR148 pKa = 11.84WHH150 pKa = 6.6KK151 pKa = 10.6VAGLADD157 pKa = 3.97YY158 pKa = 10.29NGCYY162 pKa = 9.76YY163 pKa = 10.76DD164 pKa = 4.26EE165 pKa = 4.84EE166 pKa = 5.62NGDD169 pKa = 3.22RR170 pKa = 11.84VYY172 pKa = 11.12FRR174 pKa = 11.84LFEE177 pKa = 5.09KK178 pKa = 10.73DD179 pKa = 2.75AAIYY183 pKa = 9.79GRR185 pKa = 11.84SGQWTVKK192 pKa = 10.37YY193 pKa = 10.02KK194 pKa = 9.76NTVISAPVTSSTRR207 pKa = 11.84PSDD210 pKa = 3.38WYY212 pKa = 10.96SGEE215 pKa = 4.57AGDD218 pKa = 4.42TSTSNTTTPEE228 pKa = 3.93EE229 pKa = 3.97KK230 pKa = 10.19RR231 pKa = 11.84EE232 pKa = 3.97TRR234 pKa = 11.84RR235 pKa = 11.84PHH237 pKa = 4.69QQISTEE243 pKa = 3.96ATGPGSTSPSSTTHH257 pKa = 5.2LPGRR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84RR266 pKa = 11.84SEE268 pKa = 3.78QGEE271 pKa = 4.17HH272 pKa = 6.8SSTTRR277 pKa = 11.84AKK279 pKa = 10.3RR280 pKa = 11.84RR281 pKa = 11.84RR282 pKa = 11.84TPSPGSAPSPEE293 pKa = 4.2EE294 pKa = 3.47VGRR297 pKa = 11.84VHH299 pKa = 7.8RR300 pKa = 11.84SVEE303 pKa = 3.93KK304 pKa = 10.77HH305 pKa = 4.81GLTRR309 pKa = 11.84LGRR312 pKa = 11.84IQAEE316 pKa = 4.02ARR318 pKa = 11.84DD319 pKa = 3.71PAVIIVKK326 pKa = 10.28GYY328 pKa = 11.27ANSLKK333 pKa = 10.02CWRR336 pKa = 11.84RR337 pKa = 11.84RR338 pKa = 11.84IWLKK342 pKa = 10.28HH343 pKa = 5.14RR344 pKa = 11.84SMYY347 pKa = 10.36KK348 pKa = 10.4DD349 pKa = 2.87SSTVFNWVGDD359 pKa = 4.08HH360 pKa = 6.85NSNKK364 pKa = 8.04HH365 pKa = 4.31TKK367 pKa = 10.04SRR369 pKa = 11.84ILLAFHH375 pKa = 6.17STEE378 pKa = 3.66QRR380 pKa = 11.84QLFLSSVPLPKK391 pKa = 10.3GSCMSLGNLDD401 pKa = 3.79SLL403 pKa = 4.83
MM1 pKa = 7.11EE2 pKa = 4.22TRR4 pKa = 11.84EE5 pKa = 4.04TLTARR10 pKa = 11.84FDD12 pKa = 3.73ALQEE16 pKa = 4.12QIMTLYY22 pKa = 10.52EE23 pKa = 4.0RR24 pKa = 11.84GAADD28 pKa = 3.94LQTQILHH35 pKa = 6.35WDD37 pKa = 4.09LVRR40 pKa = 11.84KK41 pKa = 9.92EE42 pKa = 4.11NVLLYY47 pKa = 10.06HH48 pKa = 6.09SRR50 pKa = 11.84KK51 pKa = 9.17QGFMSLGLQPTPALQVSEE69 pKa = 4.32YY70 pKa = 9.3RR71 pKa = 11.84AKK73 pKa = 10.59EE74 pKa = 4.11AIQMGILLNSLAKK87 pKa = 10.04SQYY90 pKa = 10.97ANEE93 pKa = 4.12RR94 pKa = 11.84WTLSDD99 pKa = 3.36TSAQLLLTEE108 pKa = 5.09PKK110 pKa = 10.2YY111 pKa = 10.75CFKK114 pKa = 10.83KK115 pKa = 10.19GAYY118 pKa = 8.05QVEE121 pKa = 4.98VYY123 pKa = 10.58FDD125 pKa = 3.55NDD127 pKa = 3.23EE128 pKa = 5.38ANMFPYY134 pKa = 10.43PNWNYY139 pKa = 10.2IYY141 pKa = 10.67YY142 pKa = 9.82QDD144 pKa = 5.05EE145 pKa = 4.12EE146 pKa = 4.51EE147 pKa = 4.37RR148 pKa = 11.84WHH150 pKa = 6.6KK151 pKa = 10.6VAGLADD157 pKa = 3.97YY158 pKa = 10.29NGCYY162 pKa = 9.76YY163 pKa = 10.76DD164 pKa = 4.26EE165 pKa = 4.84EE166 pKa = 5.62NGDD169 pKa = 3.22RR170 pKa = 11.84VYY172 pKa = 11.12FRR174 pKa = 11.84LFEE177 pKa = 5.09KK178 pKa = 10.73DD179 pKa = 2.75AAIYY183 pKa = 9.79GRR185 pKa = 11.84SGQWTVKK192 pKa = 10.37YY193 pKa = 10.02KK194 pKa = 9.76NTVISAPVTSSTRR207 pKa = 11.84PSDD210 pKa = 3.38WYY212 pKa = 10.96SGEE215 pKa = 4.57AGDD218 pKa = 4.42TSTSNTTTPEE228 pKa = 3.93EE229 pKa = 3.97KK230 pKa = 10.19RR231 pKa = 11.84EE232 pKa = 3.97TRR234 pKa = 11.84RR235 pKa = 11.84PHH237 pKa = 4.69QQISTEE243 pKa = 3.96ATGPGSTSPSSTTHH257 pKa = 5.2LPGRR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84RR266 pKa = 11.84SEE268 pKa = 3.78QGEE271 pKa = 4.17HH272 pKa = 6.8SSTTRR277 pKa = 11.84AKK279 pKa = 10.3RR280 pKa = 11.84RR281 pKa = 11.84RR282 pKa = 11.84TPSPGSAPSPEE293 pKa = 4.2EE294 pKa = 3.47VGRR297 pKa = 11.84VHH299 pKa = 7.8RR300 pKa = 11.84SVEE303 pKa = 3.93KK304 pKa = 10.77HH305 pKa = 4.81GLTRR309 pKa = 11.84LGRR312 pKa = 11.84IQAEE316 pKa = 4.02ARR318 pKa = 11.84DD319 pKa = 3.71PAVIIVKK326 pKa = 10.28GYY328 pKa = 11.27ANSLKK333 pKa = 10.02CWRR336 pKa = 11.84RR337 pKa = 11.84RR338 pKa = 11.84IWLKK342 pKa = 10.28HH343 pKa = 5.14RR344 pKa = 11.84SMYY347 pKa = 10.36KK348 pKa = 10.4DD349 pKa = 2.87SSTVFNWVGDD359 pKa = 4.08HH360 pKa = 6.85NSNKK364 pKa = 8.04HH365 pKa = 4.31TKK367 pKa = 10.04SRR369 pKa = 11.84ILLAFHH375 pKa = 6.17STEE378 pKa = 3.66QRR380 pKa = 11.84QLFLSSVPLPKK391 pKa = 10.3GSCMSLGNLDD401 pKa = 3.79SLL403 pKa = 4.83
Molecular weight: 46.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2400 |
96 |
609 |
342.9 |
38.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.5 ± 0.577 | 1.917 ± 0.713 |
6.875 ± 0.531 | 6.208 ± 0.296 |
4.125 ± 0.558 | 6.208 ± 0.851 |
1.708 ± 0.245 | 6.042 ± 0.819 |
5.625 ± 1.05 | 9.083 ± 0.695 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.708 ± 0.483 | 4.792 ± 1.035 |
6.5 ± 1.442 | 4.208 ± 0.376 |
5.458 ± 0.715 | 7.125 ± 0.85 |
6.25 ± 0.568 | 5.708 ± 0.459 |
1.25 ± 0.32 | 3.708 ± 0.293 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
