
Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Prevotellaceae; Prevotella; Prevotella ruminicola
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
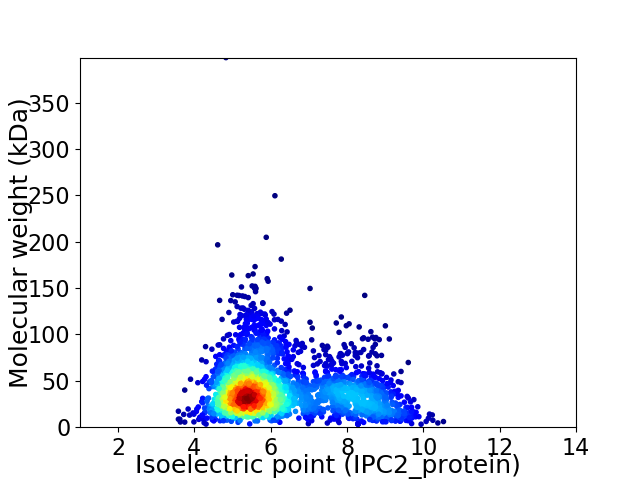
Virtual 2D-PAGE plot for 2761 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
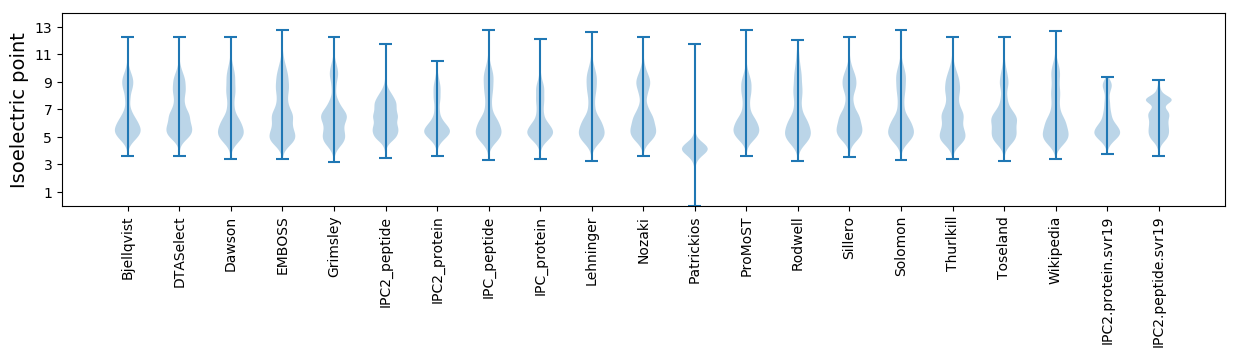
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D5EXR2|D5EXR2_PRER2 HipA domain protein OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=PRU_0523 PE=3 SV=1
MM1 pKa = 7.46KK2 pKa = 10.54NIIKK6 pKa = 10.52SSLLLLCSVAFLAACSDD23 pKa = 4.41DD24 pKa = 4.31NDD26 pKa = 5.49SNPTLQNPSTFTLNEE41 pKa = 3.86PGYY44 pKa = 11.1ASLIVDD50 pKa = 4.71LATSEE55 pKa = 5.0GIPFTWSQPDD65 pKa = 3.26YY66 pKa = 9.65GFPVAAEE73 pKa = 4.04YY74 pKa = 10.39QLQVSLDD81 pKa = 3.79GNFTTDD87 pKa = 3.37LADD90 pKa = 3.46VEE92 pKa = 4.81EE93 pKa = 5.02GNEE96 pKa = 3.96TTANFATLASYY107 pKa = 11.39YY108 pKa = 9.67NVPSGSIIPDD118 pKa = 3.36QLSNAINAMSGWDD131 pKa = 3.89GEE133 pKa = 4.79TVPQVATVYY142 pKa = 10.44VRR144 pKa = 11.84ATATTAGAPVIYY156 pKa = 10.52SNVVQIKK163 pKa = 8.61VVPNLVVAPSYY174 pKa = 11.31KK175 pKa = 10.3EE176 pKa = 3.73FIYY179 pKa = 10.42EE180 pKa = 3.94IGNEE184 pKa = 4.35SGWGTSYY191 pKa = 11.77AMRR194 pKa = 11.84SPDD197 pKa = 2.97QDD199 pKa = 3.67GIYY202 pKa = 10.76VSYY205 pKa = 10.97NWLDD209 pKa = 3.35GAFKK213 pKa = 10.48FKK215 pKa = 10.66PNEE218 pKa = 3.98NSWDD222 pKa = 3.87GDD224 pKa = 3.14WGQDD228 pKa = 3.4PNGDD232 pKa = 3.49FGTLVVDD239 pKa = 5.58GEE241 pKa = 4.37EE242 pKa = 4.53DD243 pKa = 3.63CNKK246 pKa = 10.34ADD248 pKa = 5.03GSFPDD253 pKa = 3.7NVQPAGFYY261 pKa = 10.02KK262 pKa = 10.4IQVSLVDD269 pKa = 4.06MTWSITPITNVSIIGGFNDD288 pKa = 3.35WAGDD292 pKa = 4.4VEE294 pKa = 4.72MTWNADD300 pKa = 3.31EE301 pKa = 5.05HH302 pKa = 6.79CWEE305 pKa = 4.41TTTSAVSGEE314 pKa = 4.05FKK316 pKa = 10.66FRR318 pKa = 11.84ANHH321 pKa = 5.77AWDD324 pKa = 4.06INWGGDD330 pKa = 3.32SFDD333 pKa = 4.21EE334 pKa = 4.82LKK336 pKa = 10.56QDD338 pKa = 4.04GANLSIDD345 pKa = 3.0AGTYY349 pKa = 10.04KK350 pKa = 10.53FQLYY354 pKa = 8.26LTYY357 pKa = 10.81DD358 pKa = 3.54GGSKK362 pKa = 10.41VVITAQQ368 pKa = 3.04
MM1 pKa = 7.46KK2 pKa = 10.54NIIKK6 pKa = 10.52SSLLLLCSVAFLAACSDD23 pKa = 4.41DD24 pKa = 4.31NDD26 pKa = 5.49SNPTLQNPSTFTLNEE41 pKa = 3.86PGYY44 pKa = 11.1ASLIVDD50 pKa = 4.71LATSEE55 pKa = 5.0GIPFTWSQPDD65 pKa = 3.26YY66 pKa = 9.65GFPVAAEE73 pKa = 4.04YY74 pKa = 10.39QLQVSLDD81 pKa = 3.79GNFTTDD87 pKa = 3.37LADD90 pKa = 3.46VEE92 pKa = 4.81EE93 pKa = 5.02GNEE96 pKa = 3.96TTANFATLASYY107 pKa = 11.39YY108 pKa = 9.67NVPSGSIIPDD118 pKa = 3.36QLSNAINAMSGWDD131 pKa = 3.89GEE133 pKa = 4.79TVPQVATVYY142 pKa = 10.44VRR144 pKa = 11.84ATATTAGAPVIYY156 pKa = 10.52SNVVQIKK163 pKa = 8.61VVPNLVVAPSYY174 pKa = 11.31KK175 pKa = 10.3EE176 pKa = 3.73FIYY179 pKa = 10.42EE180 pKa = 3.94IGNEE184 pKa = 4.35SGWGTSYY191 pKa = 11.77AMRR194 pKa = 11.84SPDD197 pKa = 2.97QDD199 pKa = 3.67GIYY202 pKa = 10.76VSYY205 pKa = 10.97NWLDD209 pKa = 3.35GAFKK213 pKa = 10.48FKK215 pKa = 10.66PNEE218 pKa = 3.98NSWDD222 pKa = 3.87GDD224 pKa = 3.14WGQDD228 pKa = 3.4PNGDD232 pKa = 3.49FGTLVVDD239 pKa = 5.58GEE241 pKa = 4.37EE242 pKa = 4.53DD243 pKa = 3.63CNKK246 pKa = 10.34ADD248 pKa = 5.03GSFPDD253 pKa = 3.7NVQPAGFYY261 pKa = 10.02KK262 pKa = 10.4IQVSLVDD269 pKa = 4.06MTWSITPITNVSIIGGFNDD288 pKa = 3.35WAGDD292 pKa = 4.4VEE294 pKa = 4.72MTWNADD300 pKa = 3.31EE301 pKa = 5.05HH302 pKa = 6.79CWEE305 pKa = 4.41TTTSAVSGEE314 pKa = 4.05FKK316 pKa = 10.66FRR318 pKa = 11.84ANHH321 pKa = 5.77AWDD324 pKa = 4.06INWGGDD330 pKa = 3.32SFDD333 pKa = 4.21EE334 pKa = 4.82LKK336 pKa = 10.56QDD338 pKa = 4.04GANLSIDD345 pKa = 3.0AGTYY349 pKa = 10.04KK350 pKa = 10.53FQLYY354 pKa = 8.26LTYY357 pKa = 10.81DD358 pKa = 3.54GGSKK362 pKa = 10.41VVITAQQ368 pKa = 3.04
Molecular weight: 40.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D5EYD7|D5EYD7_PRER2 30S ribosomal protein S18 OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=rpsR PE=3 SV=1
MM1 pKa = 7.02MALPAGSMAAANAGANPMGAPMGNPMANPMGNPMANPMGNPMGSPAGNPGVGAAVRR57 pKa = 11.84PGVNISVTPSASVSRR72 pKa = 11.84AANVPPAQQQQPVSPRR88 pKa = 11.84TSNAQNIAASISVGPAISIAQAGAGRR114 pKa = 11.84SSMPMMTLVNEE125 pKa = 4.33SLNIRR130 pKa = 11.84ITAMNGAIIGRR141 pKa = 11.84RR142 pKa = 11.84KK143 pKa = 10.03GPYY146 pKa = 9.0VQFFEE151 pKa = 4.41QNSYY155 pKa = 10.87VSGIHH160 pKa = 6.37AQLKK164 pKa = 8.05YY165 pKa = 10.7KK166 pKa = 10.63SGAGWFVIDD175 pKa = 4.2MNSSNGTRR183 pKa = 11.84INQAPIQPEE192 pKa = 4.18TEE194 pKa = 4.08NPLKK198 pKa = 11.09NGDD201 pKa = 3.82VLSIANVNLQVVIRR215 pKa = 4.31
MM1 pKa = 7.02MALPAGSMAAANAGANPMGAPMGNPMANPMGNPMANPMGNPMGSPAGNPGVGAAVRR57 pKa = 11.84PGVNISVTPSASVSRR72 pKa = 11.84AANVPPAQQQQPVSPRR88 pKa = 11.84TSNAQNIAASISVGPAISIAQAGAGRR114 pKa = 11.84SSMPMMTLVNEE125 pKa = 4.33SLNIRR130 pKa = 11.84ITAMNGAIIGRR141 pKa = 11.84RR142 pKa = 11.84KK143 pKa = 10.03GPYY146 pKa = 9.0VQFFEE151 pKa = 4.41QNSYY155 pKa = 10.87VSGIHH160 pKa = 6.37AQLKK164 pKa = 8.05YY165 pKa = 10.7KK166 pKa = 10.63SGAGWFVIDD175 pKa = 4.2MNSSNGTRR183 pKa = 11.84INQAPIQPEE192 pKa = 4.18TEE194 pKa = 4.08NPLKK198 pKa = 11.09NGDD201 pKa = 3.82VLSIANVNLQVVIRR215 pKa = 4.31
Molecular weight: 22.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1065923 |
30 |
3655 |
386.1 |
43.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.596 ± 0.047 | 1.338 ± 0.017 |
5.956 ± 0.029 | 6.265 ± 0.041 |
4.275 ± 0.025 | 6.958 ± 0.037 |
2.08 ± 0.023 | 6.543 ± 0.04 |
6.455 ± 0.047 | 8.748 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.997 ± 0.02 | 4.979 ± 0.035 |
3.724 ± 0.023 | 3.966 ± 0.028 |
4.579 ± 0.035 | 5.602 ± 0.032 |
5.696 ± 0.037 | 6.491 ± 0.033 |
1.389 ± 0.02 | 4.364 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
