
Firmicutes bacterium CAG:882
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; environmental samples
Average proteome isoelectric point is 5.86
Get precalculated fractions of proteins
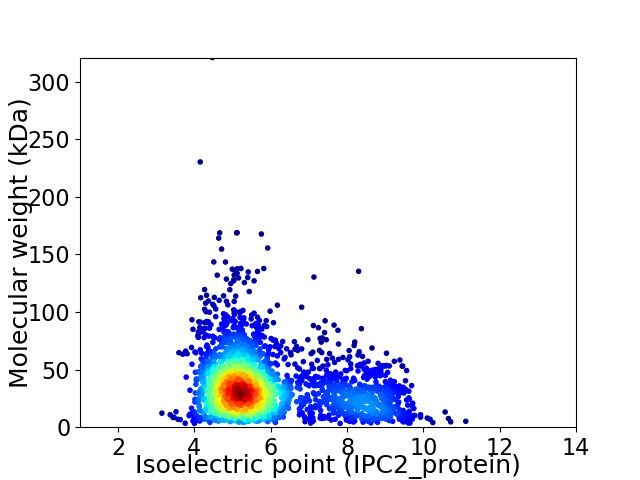
Virtual 2D-PAGE plot for 2572 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
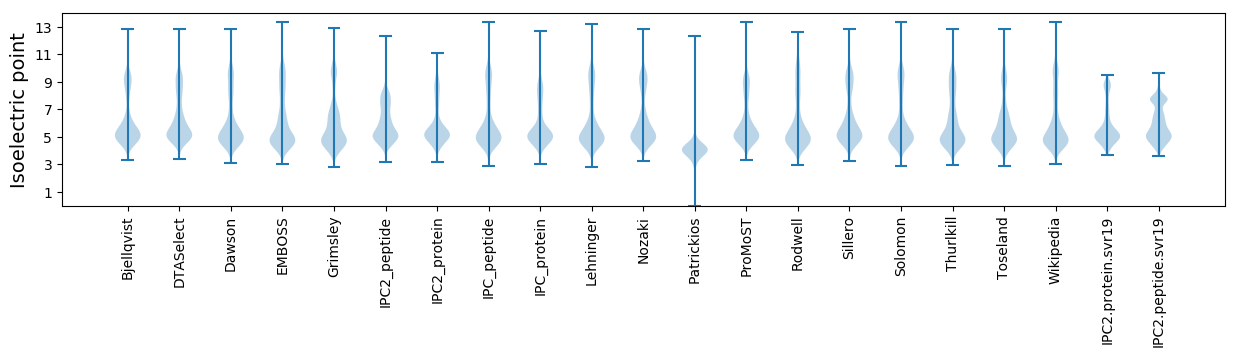
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7BL48|R7BL48_9FIRM ATP synthase subunit c OS=Firmicutes bacterium CAG:882 OX=1262991 GN=atpE PE=3 SV=1
MM1 pKa = 7.79RR2 pKa = 11.84KK3 pKa = 9.08FKK5 pKa = 10.7KK6 pKa = 10.52VLASLIAGTLVLSNAVPVLAAQDD29 pKa = 3.93ASALPDD35 pKa = 3.12GTAYY39 pKa = 11.2LNINNSDD46 pKa = 3.4WQPFDD51 pKa = 4.66AEE53 pKa = 4.32WVNAEE58 pKa = 3.75ITGDD62 pKa = 3.4GSYY65 pKa = 9.44TVSMTAAEE73 pKa = 4.26AVDD76 pKa = 4.68LGAFNALEE84 pKa = 4.19VVNGEE89 pKa = 4.25EE90 pKa = 4.13VLGTGAVITVDD101 pKa = 3.76SIEE104 pKa = 4.38LNGEE108 pKa = 4.08EE109 pKa = 5.18IEE111 pKa = 4.44LQGSSYY117 pKa = 10.05TCSADD122 pKa = 3.2GLGVTTRR129 pKa = 11.84VNLYY133 pKa = 10.34NEE135 pKa = 4.4WNTPDD140 pKa = 3.27ATATAGDD147 pKa = 4.81DD148 pKa = 3.21KK149 pKa = 11.5HH150 pKa = 7.47ADD152 pKa = 3.43CRR154 pKa = 11.84VAEE157 pKa = 4.45GNVADD162 pKa = 4.59ATACLWTAEE171 pKa = 4.1QLTGVTSIVVNFTVSGFGTAGGPAEE196 pKa = 4.54AADD199 pKa = 3.81NSDD202 pKa = 3.29AATEE206 pKa = 4.19AVAHH210 pKa = 5.34LTINNEE216 pKa = 3.29NWGEE220 pKa = 4.04YY221 pKa = 7.62EE222 pKa = 4.27AEE224 pKa = 4.19YY225 pKa = 11.33VDD227 pKa = 4.28ATITGDD233 pKa = 3.23GTYY236 pKa = 9.15TVSMTAAEE244 pKa = 4.12AQNLGAYY251 pKa = 8.33NALQVPNGEE260 pKa = 4.74AIMGTACVLTIDD272 pKa = 4.13SIKK275 pKa = 10.81INGEE279 pKa = 3.98EE280 pKa = 4.01ITLQGPSYY288 pKa = 10.0TCSADD293 pKa = 3.18GAGVDD298 pKa = 3.69TRR300 pKa = 11.84VNIYY304 pKa = 10.7NEE306 pKa = 4.18YY307 pKa = 10.4NDD309 pKa = 4.27PDD311 pKa = 3.62ATATAGDD318 pKa = 4.8DD319 pKa = 3.3NHH321 pKa = 7.14LDD323 pKa = 3.62QRR325 pKa = 11.84IADD328 pKa = 4.13GNLADD333 pKa = 3.57ATARR337 pKa = 11.84LLSTDD342 pKa = 5.08DD343 pKa = 3.45IADD346 pKa = 3.73VQSVEE351 pKa = 4.11VTFTVSDD358 pKa = 3.75YY359 pKa = 11.75GKK361 pKa = 8.92MAEE364 pKa = 4.25AGEE367 pKa = 4.13AAGNDD372 pKa = 3.75GAFDD376 pKa = 4.45PNGEE380 pKa = 4.18YY381 pKa = 10.47NVYY384 pKa = 10.65FGVQTAAYY392 pKa = 9.4SFRR395 pKa = 11.84NAWNDD400 pKa = 3.23SYY402 pKa = 12.13GLGTPEE408 pKa = 4.23FDD410 pKa = 5.96QITGWNGNDD419 pKa = 2.85EE420 pKa = 4.51VVRR423 pKa = 11.84TGTITDD429 pKa = 3.69AVIKK433 pKa = 10.93GNGTYY438 pKa = 9.16TVSLTGVDD446 pKa = 4.27FADD449 pKa = 4.84DD450 pKa = 4.43AEE452 pKa = 4.39TLNLLFLSTDD462 pKa = 3.05IPLDD466 pKa = 3.63SDD468 pKa = 4.67AKK470 pKa = 10.03ITDD473 pKa = 3.33ITVKK477 pKa = 10.1MGGKK481 pKa = 9.17KK482 pKa = 10.44VYY484 pKa = 10.33DD485 pKa = 3.93FEE487 pKa = 5.68EE488 pKa = 5.27SYY490 pKa = 11.31QDD492 pKa = 4.52PDD494 pKa = 3.35TKK496 pKa = 11.11DD497 pKa = 3.35YY498 pKa = 11.34IKK500 pKa = 10.04TLLINTYY507 pKa = 10.07NDD509 pKa = 3.04AFNGEE514 pKa = 4.11NEE516 pKa = 4.05FAYY519 pKa = 9.63TLSAGMDD526 pKa = 3.2IEE528 pKa = 4.59VTFTVSGFAYY538 pKa = 9.94DD539 pKa = 4.17NPDD542 pKa = 3.45AVPSGDD548 pKa = 3.26TSATEE553 pKa = 3.97TTQAAEE559 pKa = 4.17TTKK562 pKa = 10.63AASEE566 pKa = 4.19ATTTASAEE574 pKa = 3.96ASQGGVSVGVIIAIVAAVVVVAAIVVVAVKK604 pKa = 10.27KK605 pKa = 10.74KK606 pKa = 10.17KK607 pKa = 10.34
MM1 pKa = 7.79RR2 pKa = 11.84KK3 pKa = 9.08FKK5 pKa = 10.7KK6 pKa = 10.52VLASLIAGTLVLSNAVPVLAAQDD29 pKa = 3.93ASALPDD35 pKa = 3.12GTAYY39 pKa = 11.2LNINNSDD46 pKa = 3.4WQPFDD51 pKa = 4.66AEE53 pKa = 4.32WVNAEE58 pKa = 3.75ITGDD62 pKa = 3.4GSYY65 pKa = 9.44TVSMTAAEE73 pKa = 4.26AVDD76 pKa = 4.68LGAFNALEE84 pKa = 4.19VVNGEE89 pKa = 4.25EE90 pKa = 4.13VLGTGAVITVDD101 pKa = 3.76SIEE104 pKa = 4.38LNGEE108 pKa = 4.08EE109 pKa = 5.18IEE111 pKa = 4.44LQGSSYY117 pKa = 10.05TCSADD122 pKa = 3.2GLGVTTRR129 pKa = 11.84VNLYY133 pKa = 10.34NEE135 pKa = 4.4WNTPDD140 pKa = 3.27ATATAGDD147 pKa = 4.81DD148 pKa = 3.21KK149 pKa = 11.5HH150 pKa = 7.47ADD152 pKa = 3.43CRR154 pKa = 11.84VAEE157 pKa = 4.45GNVADD162 pKa = 4.59ATACLWTAEE171 pKa = 4.1QLTGVTSIVVNFTVSGFGTAGGPAEE196 pKa = 4.54AADD199 pKa = 3.81NSDD202 pKa = 3.29AATEE206 pKa = 4.19AVAHH210 pKa = 5.34LTINNEE216 pKa = 3.29NWGEE220 pKa = 4.04YY221 pKa = 7.62EE222 pKa = 4.27AEE224 pKa = 4.19YY225 pKa = 11.33VDD227 pKa = 4.28ATITGDD233 pKa = 3.23GTYY236 pKa = 9.15TVSMTAAEE244 pKa = 4.12AQNLGAYY251 pKa = 8.33NALQVPNGEE260 pKa = 4.74AIMGTACVLTIDD272 pKa = 4.13SIKK275 pKa = 10.81INGEE279 pKa = 3.98EE280 pKa = 4.01ITLQGPSYY288 pKa = 10.0TCSADD293 pKa = 3.18GAGVDD298 pKa = 3.69TRR300 pKa = 11.84VNIYY304 pKa = 10.7NEE306 pKa = 4.18YY307 pKa = 10.4NDD309 pKa = 4.27PDD311 pKa = 3.62ATATAGDD318 pKa = 4.8DD319 pKa = 3.3NHH321 pKa = 7.14LDD323 pKa = 3.62QRR325 pKa = 11.84IADD328 pKa = 4.13GNLADD333 pKa = 3.57ATARR337 pKa = 11.84LLSTDD342 pKa = 5.08DD343 pKa = 3.45IADD346 pKa = 3.73VQSVEE351 pKa = 4.11VTFTVSDD358 pKa = 3.75YY359 pKa = 11.75GKK361 pKa = 8.92MAEE364 pKa = 4.25AGEE367 pKa = 4.13AAGNDD372 pKa = 3.75GAFDD376 pKa = 4.45PNGEE380 pKa = 4.18YY381 pKa = 10.47NVYY384 pKa = 10.65FGVQTAAYY392 pKa = 9.4SFRR395 pKa = 11.84NAWNDD400 pKa = 3.23SYY402 pKa = 12.13GLGTPEE408 pKa = 4.23FDD410 pKa = 5.96QITGWNGNDD419 pKa = 2.85EE420 pKa = 4.51VVRR423 pKa = 11.84TGTITDD429 pKa = 3.69AVIKK433 pKa = 10.93GNGTYY438 pKa = 9.16TVSLTGVDD446 pKa = 4.27FADD449 pKa = 4.84DD450 pKa = 4.43AEE452 pKa = 4.39TLNLLFLSTDD462 pKa = 3.05IPLDD466 pKa = 3.63SDD468 pKa = 4.67AKK470 pKa = 10.03ITDD473 pKa = 3.33ITVKK477 pKa = 10.1MGGKK481 pKa = 9.17KK482 pKa = 10.44VYY484 pKa = 10.33DD485 pKa = 3.93FEE487 pKa = 5.68EE488 pKa = 5.27SYY490 pKa = 11.31QDD492 pKa = 4.52PDD494 pKa = 3.35TKK496 pKa = 11.11DD497 pKa = 3.35YY498 pKa = 11.34IKK500 pKa = 10.04TLLINTYY507 pKa = 10.07NDD509 pKa = 3.04AFNGEE514 pKa = 4.11NEE516 pKa = 4.05FAYY519 pKa = 9.63TLSAGMDD526 pKa = 3.2IEE528 pKa = 4.59VTFTVSGFAYY538 pKa = 9.94DD539 pKa = 4.17NPDD542 pKa = 3.45AVPSGDD548 pKa = 3.26TSATEE553 pKa = 3.97TTQAAEE559 pKa = 4.17TTKK562 pKa = 10.63AASEE566 pKa = 4.19ATTTASAEE574 pKa = 3.96ASQGGVSVGVIIAIVAAVVVVAAIVVVAVKK604 pKa = 10.27KK605 pKa = 10.74KK606 pKa = 10.17KK607 pKa = 10.34
Molecular weight: 63.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7BCH2|R7BCH2_9FIRM Extracellular solute-binding protein family 1 OS=Firmicutes bacterium CAG:882 OX=1262991 GN=BN803_01148 PE=4 SV=1
MM1 pKa = 7.33KK2 pKa = 9.54QTFHH6 pKa = 6.38PKK8 pKa = 9.07KK9 pKa = 8.04RR10 pKa = 11.84QRR12 pKa = 11.84NKK14 pKa = 8.21VHH16 pKa = 6.58GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.98VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.03GRR39 pKa = 11.84KK40 pKa = 8.97SLTVV44 pKa = 3.12
MM1 pKa = 7.33KK2 pKa = 9.54QTFHH6 pKa = 6.38PKK8 pKa = 9.07KK9 pKa = 8.04RR10 pKa = 11.84QRR12 pKa = 11.84NKK14 pKa = 8.21VHH16 pKa = 6.58GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.98VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.03GRR39 pKa = 11.84KK40 pKa = 8.97SLTVV44 pKa = 3.12
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
845954 |
29 |
2973 |
328.9 |
36.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.614 ± 0.045 | 1.509 ± 0.019 |
6.284 ± 0.042 | 7.217 ± 0.048 |
4.07 ± 0.033 | 7.061 ± 0.044 |
1.625 ± 0.02 | 7.568 ± 0.045 |
6.674 ± 0.045 | 8.442 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.134 ± 0.024 | 4.883 ± 0.03 |
2.916 ± 0.026 | 2.607 ± 0.025 |
4.357 ± 0.038 | 6.006 ± 0.042 |
5.339 ± 0.046 | 7.228 ± 0.04 |
0.85 ± 0.017 | 4.613 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
