
Lactobacillus phage Ldl1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Tybeckvirinae; Lidleunavirus; Lactobacillus virus Ldl1
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
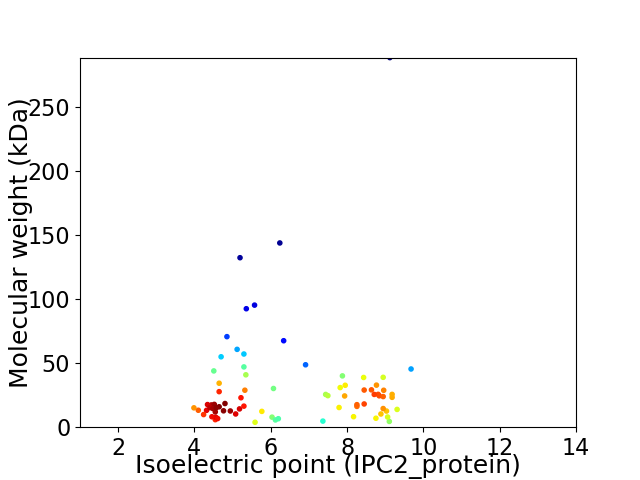
Virtual 2D-PAGE plot for 79 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A7DMT0|A0A0A7DMT0_9CAUD HNH endonuclease OS=Lactobacillus phage Ldl1 OX=1552735 GN=LDL_055 PE=4 SV=1
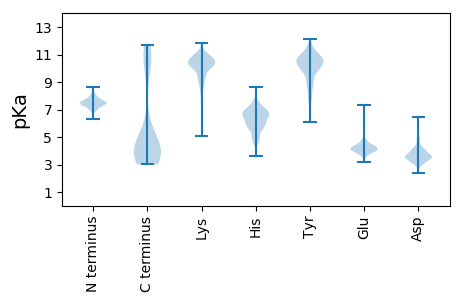
MM1 pKa = 7.43QSRR4 pKa = 11.84FKK6 pKa = 10.43IAKK9 pKa = 8.28EE10 pKa = 3.71FNKK13 pKa = 10.17IIAKK17 pKa = 10.06ACEE20 pKa = 4.08DD21 pKa = 3.51QPLEE25 pKa = 4.39DD26 pKa = 4.31FLEE29 pKa = 4.32EE30 pKa = 4.68EE31 pKa = 4.39IEE33 pKa = 4.14EE34 pKa = 4.54DD35 pKa = 4.74CSGLDD40 pKa = 4.04FDD42 pKa = 6.28DD43 pKa = 4.96AAQLYY48 pKa = 10.52DD49 pKa = 3.97EE50 pKa = 5.23YY51 pKa = 11.15RR52 pKa = 11.84DD53 pKa = 3.75EE54 pKa = 4.92CIEE57 pKa = 3.72WFNYY61 pKa = 9.19FFEE64 pKa = 4.62KK65 pKa = 10.21TCMSPQEE72 pKa = 4.08FCVLCDD78 pKa = 3.1WDD80 pKa = 4.06YY81 pKa = 12.13VLDD84 pKa = 4.03EE85 pKa = 5.09EE86 pKa = 4.52NDD88 pKa = 3.35KK89 pKa = 11.05FLVMDD94 pKa = 3.81TDD96 pKa = 3.72KK97 pKa = 11.75NKK99 pKa = 10.14FLLVTSMFEE108 pKa = 4.1HH109 pKa = 6.29YY110 pKa = 10.37CIEE113 pKa = 3.98IFNNLDD119 pKa = 2.93VHH121 pKa = 6.97NIDD124 pKa = 4.25KK125 pKa = 11.06
MM1 pKa = 7.43QSRR4 pKa = 11.84FKK6 pKa = 10.43IAKK9 pKa = 8.28EE10 pKa = 3.71FNKK13 pKa = 10.17IIAKK17 pKa = 10.06ACEE20 pKa = 4.08DD21 pKa = 3.51QPLEE25 pKa = 4.39DD26 pKa = 4.31FLEE29 pKa = 4.32EE30 pKa = 4.68EE31 pKa = 4.39IEE33 pKa = 4.14EE34 pKa = 4.54DD35 pKa = 4.74CSGLDD40 pKa = 4.04FDD42 pKa = 6.28DD43 pKa = 4.96AAQLYY48 pKa = 10.52DD49 pKa = 3.97EE50 pKa = 5.23YY51 pKa = 11.15RR52 pKa = 11.84DD53 pKa = 3.75EE54 pKa = 4.92CIEE57 pKa = 3.72WFNYY61 pKa = 9.19FFEE64 pKa = 4.62KK65 pKa = 10.21TCMSPQEE72 pKa = 4.08FCVLCDD78 pKa = 3.1WDD80 pKa = 4.06YY81 pKa = 12.13VLDD84 pKa = 4.03EE85 pKa = 5.09EE86 pKa = 4.52NDD88 pKa = 3.35KK89 pKa = 11.05FLVMDD94 pKa = 3.81TDD96 pKa = 3.72KK97 pKa = 11.75NKK99 pKa = 10.14FLLVTSMFEE108 pKa = 4.1HH109 pKa = 6.29YY110 pKa = 10.37CIEE113 pKa = 3.98IFNNLDD119 pKa = 2.93VHH121 pKa = 6.97NIDD124 pKa = 4.25KK125 pKa = 11.06
Molecular weight: 15.1 kDa
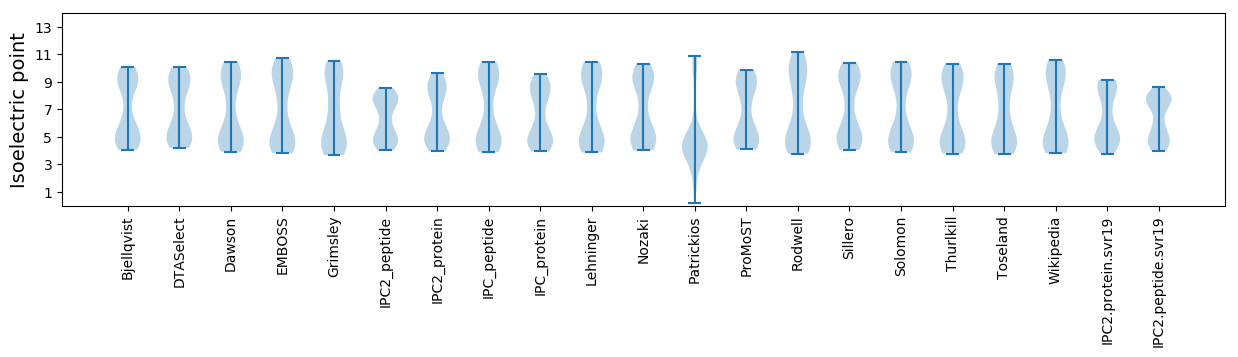
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A7DML2|A0A0A7DML2_9CAUD HNH endonuclease OS=Lactobacillus phage Ldl1 OX=1552735 GN=LDL_039 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 10.4KK3 pKa = 10.3KK4 pKa = 10.31IIPILLSSATVLGLMFANKK23 pKa = 9.43ISAATRR29 pKa = 11.84EE30 pKa = 4.11EE31 pKa = 4.29GVDD34 pKa = 3.38VSSYY38 pKa = 10.36QGSSSSYY45 pKa = 10.72FKK47 pKa = 10.23TLKK50 pKa = 10.79NKK52 pKa = 8.8GAKK55 pKa = 9.21FVIVKK60 pKa = 10.29VGGSGGGEE68 pKa = 4.17GYY70 pKa = 10.53HH71 pKa = 5.46YY72 pKa = 10.18QNPSASMQLANAKK85 pKa = 10.23VNGLAVGAYY94 pKa = 8.37FWAEE98 pKa = 4.15CGSSSSEE105 pKa = 3.86ALRR108 pKa = 11.84MAKK111 pKa = 10.24LAVSDD116 pKa = 4.17AKK118 pKa = 10.78RR119 pKa = 11.84AGLKK123 pKa = 8.66TSSVIAMDD131 pKa = 4.0YY132 pKa = 10.57EE133 pKa = 5.14AGAYY137 pKa = 9.6ASQKK141 pKa = 8.01TANTNAVITFMDD153 pKa = 4.8HH154 pKa = 6.14IKK156 pKa = 10.53KK157 pKa = 10.44AGYY160 pKa = 9.63KK161 pKa = 9.32PLFYY165 pKa = 10.61SGASFARR172 pKa = 11.84SNVDD176 pKa = 2.97TKK178 pKa = 11.54KK179 pKa = 10.77LVNKK183 pKa = 9.63YY184 pKa = 9.94GKK186 pKa = 9.37SALWIASYY194 pKa = 8.59KK195 pKa = 9.42TMNAQYY201 pKa = 9.87TPDD204 pKa = 3.51YY205 pKa = 10.88NYY207 pKa = 9.75FPSMDD212 pKa = 4.97GIGIWQYY219 pKa = 10.96ADD221 pKa = 2.54NWKK224 pKa = 10.25GLNVDD229 pKa = 4.08GNVQFFKK236 pKa = 10.88VISNGEE242 pKa = 4.24VTKK245 pKa = 9.63KK246 pKa = 9.32TPVKK250 pKa = 10.06QANATTNKK258 pKa = 9.83EE259 pKa = 3.91NGVKK263 pKa = 10.29KK264 pKa = 10.37FPKK267 pKa = 10.35DD268 pKa = 3.11NTYY271 pKa = 8.8TVKK274 pKa = 10.95NGDD277 pKa = 3.42SWSSIAKK284 pKa = 10.24AYY286 pKa = 10.35GIDD289 pKa = 3.7VNGLAKK295 pKa = 10.87LNGATTKK302 pKa = 10.46TMLHH306 pKa = 7.01PGQKK310 pKa = 10.12LKK312 pKa = 10.09LTGHH316 pKa = 6.29VSNVATKK323 pKa = 10.2KK324 pKa = 9.81PKK326 pKa = 9.26KK327 pKa = 7.15TTTKK331 pKa = 9.97KK332 pKa = 9.44KK333 pKa = 8.1TVKK336 pKa = 9.24KK337 pKa = 9.97HH338 pKa = 4.23SWIKK342 pKa = 9.21KK343 pKa = 8.27SGYY346 pKa = 9.33FRR348 pKa = 11.84LNTTIKK354 pKa = 10.04LRR356 pKa = 11.84SGASTSSRR364 pKa = 11.84VISTLYY370 pKa = 9.66RR371 pKa = 11.84GQVIKK376 pKa = 10.59FDD378 pKa = 3.8AYY380 pKa = 9.7KK381 pKa = 10.52ISGGYY386 pKa = 8.32VWLRR390 pKa = 11.84QKK392 pKa = 10.68RR393 pKa = 11.84SNGYY397 pKa = 9.03GSVASGVAKK406 pKa = 10.34NGKK409 pKa = 9.02RR410 pKa = 11.84VSTWGTIFF418 pKa = 3.98
MM1 pKa = 7.45KK2 pKa = 10.4KK3 pKa = 10.3KK4 pKa = 10.31IIPILLSSATVLGLMFANKK23 pKa = 9.43ISAATRR29 pKa = 11.84EE30 pKa = 4.11EE31 pKa = 4.29GVDD34 pKa = 3.38VSSYY38 pKa = 10.36QGSSSSYY45 pKa = 10.72FKK47 pKa = 10.23TLKK50 pKa = 10.79NKK52 pKa = 8.8GAKK55 pKa = 9.21FVIVKK60 pKa = 10.29VGGSGGGEE68 pKa = 4.17GYY70 pKa = 10.53HH71 pKa = 5.46YY72 pKa = 10.18QNPSASMQLANAKK85 pKa = 10.23VNGLAVGAYY94 pKa = 8.37FWAEE98 pKa = 4.15CGSSSSEE105 pKa = 3.86ALRR108 pKa = 11.84MAKK111 pKa = 10.24LAVSDD116 pKa = 4.17AKK118 pKa = 10.78RR119 pKa = 11.84AGLKK123 pKa = 8.66TSSVIAMDD131 pKa = 4.0YY132 pKa = 10.57EE133 pKa = 5.14AGAYY137 pKa = 9.6ASQKK141 pKa = 8.01TANTNAVITFMDD153 pKa = 4.8HH154 pKa = 6.14IKK156 pKa = 10.53KK157 pKa = 10.44AGYY160 pKa = 9.63KK161 pKa = 9.32PLFYY165 pKa = 10.61SGASFARR172 pKa = 11.84SNVDD176 pKa = 2.97TKK178 pKa = 11.54KK179 pKa = 10.77LVNKK183 pKa = 9.63YY184 pKa = 9.94GKK186 pKa = 9.37SALWIASYY194 pKa = 8.59KK195 pKa = 9.42TMNAQYY201 pKa = 9.87TPDD204 pKa = 3.51YY205 pKa = 10.88NYY207 pKa = 9.75FPSMDD212 pKa = 4.97GIGIWQYY219 pKa = 10.96ADD221 pKa = 2.54NWKK224 pKa = 10.25GLNVDD229 pKa = 4.08GNVQFFKK236 pKa = 10.88VISNGEE242 pKa = 4.24VTKK245 pKa = 9.63KK246 pKa = 9.32TPVKK250 pKa = 10.06QANATTNKK258 pKa = 9.83EE259 pKa = 3.91NGVKK263 pKa = 10.29KK264 pKa = 10.37FPKK267 pKa = 10.35DD268 pKa = 3.11NTYY271 pKa = 8.8TVKK274 pKa = 10.95NGDD277 pKa = 3.42SWSSIAKK284 pKa = 10.24AYY286 pKa = 10.35GIDD289 pKa = 3.7VNGLAKK295 pKa = 10.87LNGATTKK302 pKa = 10.46TMLHH306 pKa = 7.01PGQKK310 pKa = 10.12LKK312 pKa = 10.09LTGHH316 pKa = 6.29VSNVATKK323 pKa = 10.2KK324 pKa = 9.81PKK326 pKa = 9.26KK327 pKa = 7.15TTTKK331 pKa = 9.97KK332 pKa = 9.44KK333 pKa = 8.1TVKK336 pKa = 9.24KK337 pKa = 9.97HH338 pKa = 4.23SWIKK342 pKa = 9.21KK343 pKa = 8.27SGYY346 pKa = 9.33FRR348 pKa = 11.84LNTTIKK354 pKa = 10.04LRR356 pKa = 11.84SGASTSSRR364 pKa = 11.84VISTLYY370 pKa = 9.66RR371 pKa = 11.84GQVIKK376 pKa = 10.59FDD378 pKa = 3.8AYY380 pKa = 9.7KK381 pKa = 10.52ISGGYY386 pKa = 8.32VWLRR390 pKa = 11.84QKK392 pKa = 10.68RR393 pKa = 11.84SNGYY397 pKa = 9.03GSVASGVAKK406 pKa = 10.34NGKK409 pKa = 9.02RR410 pKa = 11.84VSTWGTIFF418 pKa = 3.98
Molecular weight: 45.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
21614 |
35 |
2627 |
273.6 |
31.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.389 ± 0.478 | 0.736 ± 0.12 |
6.264 ± 0.346 | 6.838 ± 0.375 |
4.071 ± 0.27 | 5.959 ± 0.24 |
1.712 ± 0.093 | 6.093 ± 0.207 |
8.999 ± 0.505 | 8.346 ± 0.271 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.017 ± 0.114 | 6.875 ± 0.171 |
2.42 ± 0.162 | 4.085 ± 0.192 |
3.9 ± 0.168 | 7.065 ± 0.305 |
6.061 ± 0.383 | 6.699 ± 0.205 |
1.189 ± 0.099 | 4.28 ± 0.211 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
