
Weissella koreensis (strain KACC 15510)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Weissella; Weissella koreensis
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
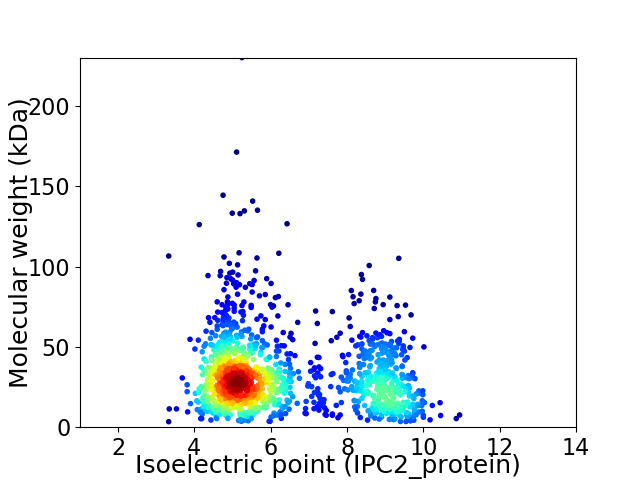
Virtual 2D-PAGE plot for 1346 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F8HXX9|F8HXX9_WEIKK PST family polysaccharide transporter OS=Weissella koreensis (strain KACC 15510) OX=1045854 GN=WKK_00395 PE=4 SV=1
MM1 pKa = 7.07QSYY4 pKa = 10.0AEE6 pKa = 4.32KK7 pKa = 10.39MLDD10 pKa = 3.68EE11 pKa = 5.11LDD13 pKa = 4.12GGQLEE18 pKa = 4.17AAKK21 pKa = 10.48KK22 pKa = 10.51SFALSLRR29 pKa = 11.84HH30 pKa = 7.01DD31 pKa = 4.66DD32 pKa = 5.28DD33 pKa = 3.98EE34 pKa = 5.23TIHH37 pKa = 6.93SLAEE41 pKa = 3.55EE42 pKa = 4.82LYY44 pKa = 11.02ALGFSTNAKK53 pKa = 9.63RR54 pKa = 11.84AYY56 pKa = 9.92QKK58 pKa = 11.32LLDD61 pKa = 4.59KK62 pKa = 11.52YY63 pKa = 10.22PDD65 pKa = 3.47EE66 pKa = 5.06DD67 pKa = 3.73VLRR70 pKa = 11.84TEE72 pKa = 4.41LAEE75 pKa = 3.94ISISEE80 pKa = 4.2DD81 pKa = 3.31QSDD84 pKa = 3.5EE85 pKa = 3.81ALAYY89 pKa = 9.09LIPIQSDD96 pKa = 3.5SEE98 pKa = 4.55AYY100 pKa = 10.63LEE102 pKa = 4.31ALLVYY107 pKa = 10.72ADD109 pKa = 5.11LYY111 pKa = 10.73QSEE114 pKa = 4.65GLLEE118 pKa = 4.09AAEE121 pKa = 4.27NKK123 pKa = 8.78LTEE126 pKa = 4.69AYY128 pKa = 10.14QLAPDD133 pKa = 3.67EE134 pKa = 4.92LVIQFALAEE143 pKa = 4.18FLFEE147 pKa = 4.62LGRR150 pKa = 11.84YY151 pKa = 8.96SEE153 pKa = 5.72AIPFYY158 pKa = 10.9RR159 pKa = 11.84NLIVQGEE166 pKa = 4.39TQISGIDD173 pKa = 3.41LVSRR177 pKa = 11.84IGVAYY182 pKa = 10.56AMLGDD187 pKa = 3.67QNRR190 pKa = 11.84ALGYY194 pKa = 10.44LEE196 pKa = 4.28QIKK199 pKa = 10.23DD200 pKa = 3.51ANLTPEE206 pKa = 3.87VRR208 pKa = 11.84FQLGMLYY215 pKa = 10.78ANDD218 pKa = 3.92EE219 pKa = 4.25EE220 pKa = 4.74TRR222 pKa = 11.84DD223 pKa = 3.45QGIEE227 pKa = 3.93AFEE230 pKa = 4.02KK231 pKa = 10.71LIEE234 pKa = 4.58IDD236 pKa = 3.29PSYY239 pKa = 11.28AGVYY243 pKa = 9.77VPLGQLYY250 pKa = 7.72EE251 pKa = 4.11QKK253 pKa = 10.8QMPKK257 pKa = 9.95DD258 pKa = 3.29ALGIYY263 pKa = 8.9EE264 pKa = 5.05AGLAVDD270 pKa = 4.1QFNEE274 pKa = 3.89AGYY277 pKa = 11.17LNATRR282 pKa = 11.84VAIQLNEE289 pKa = 3.87NDD291 pKa = 3.63QAEE294 pKa = 4.21QLYY297 pKa = 10.5QKK299 pKa = 10.41GLKK302 pKa = 9.54NLPDD306 pKa = 3.82SQNLISNYY314 pKa = 9.6SQMLVDD320 pKa = 3.76TEE322 pKa = 4.47QYY324 pKa = 9.29MEE326 pKa = 4.64QINFLNQYY334 pKa = 9.69VSADD338 pKa = 3.89DD339 pKa = 4.57EE340 pKa = 4.89FEE342 pKa = 5.03LDD344 pKa = 3.94PKK346 pKa = 10.74EE347 pKa = 4.21YY348 pKa = 10.05WNLAQSYY355 pKa = 8.45TEE357 pKa = 3.96LEE359 pKa = 4.29RR360 pKa = 11.84YY361 pKa = 9.34EE362 pKa = 4.41MADD365 pKa = 3.44QYY367 pKa = 10.14WQAAVPFFNEE377 pKa = 3.13NSIFLKK383 pKa = 10.04EE384 pKa = 3.81AIYY387 pKa = 10.13YY388 pKa = 9.66FRR390 pKa = 11.84EE391 pKa = 4.43AGNHH395 pKa = 5.26EE396 pKa = 4.52MLSDD400 pKa = 4.31LLNKK404 pKa = 9.56YY405 pKa = 9.23VQLNPEE411 pKa = 4.54DD412 pKa = 4.94FEE414 pKa = 4.39MAQMEE419 pKa = 4.85SEE421 pKa = 4.26INDD424 pKa = 3.65LL425 pKa = 4.37
MM1 pKa = 7.07QSYY4 pKa = 10.0AEE6 pKa = 4.32KK7 pKa = 10.39MLDD10 pKa = 3.68EE11 pKa = 5.11LDD13 pKa = 4.12GGQLEE18 pKa = 4.17AAKK21 pKa = 10.48KK22 pKa = 10.51SFALSLRR29 pKa = 11.84HH30 pKa = 7.01DD31 pKa = 4.66DD32 pKa = 5.28DD33 pKa = 3.98EE34 pKa = 5.23TIHH37 pKa = 6.93SLAEE41 pKa = 3.55EE42 pKa = 4.82LYY44 pKa = 11.02ALGFSTNAKK53 pKa = 9.63RR54 pKa = 11.84AYY56 pKa = 9.92QKK58 pKa = 11.32LLDD61 pKa = 4.59KK62 pKa = 11.52YY63 pKa = 10.22PDD65 pKa = 3.47EE66 pKa = 5.06DD67 pKa = 3.73VLRR70 pKa = 11.84TEE72 pKa = 4.41LAEE75 pKa = 3.94ISISEE80 pKa = 4.2DD81 pKa = 3.31QSDD84 pKa = 3.5EE85 pKa = 3.81ALAYY89 pKa = 9.09LIPIQSDD96 pKa = 3.5SEE98 pKa = 4.55AYY100 pKa = 10.63LEE102 pKa = 4.31ALLVYY107 pKa = 10.72ADD109 pKa = 5.11LYY111 pKa = 10.73QSEE114 pKa = 4.65GLLEE118 pKa = 4.09AAEE121 pKa = 4.27NKK123 pKa = 8.78LTEE126 pKa = 4.69AYY128 pKa = 10.14QLAPDD133 pKa = 3.67EE134 pKa = 4.92LVIQFALAEE143 pKa = 4.18FLFEE147 pKa = 4.62LGRR150 pKa = 11.84YY151 pKa = 8.96SEE153 pKa = 5.72AIPFYY158 pKa = 10.9RR159 pKa = 11.84NLIVQGEE166 pKa = 4.39TQISGIDD173 pKa = 3.41LVSRR177 pKa = 11.84IGVAYY182 pKa = 10.56AMLGDD187 pKa = 3.67QNRR190 pKa = 11.84ALGYY194 pKa = 10.44LEE196 pKa = 4.28QIKK199 pKa = 10.23DD200 pKa = 3.51ANLTPEE206 pKa = 3.87VRR208 pKa = 11.84FQLGMLYY215 pKa = 10.78ANDD218 pKa = 3.92EE219 pKa = 4.25EE220 pKa = 4.74TRR222 pKa = 11.84DD223 pKa = 3.45QGIEE227 pKa = 3.93AFEE230 pKa = 4.02KK231 pKa = 10.71LIEE234 pKa = 4.58IDD236 pKa = 3.29PSYY239 pKa = 11.28AGVYY243 pKa = 9.77VPLGQLYY250 pKa = 7.72EE251 pKa = 4.11QKK253 pKa = 10.8QMPKK257 pKa = 9.95DD258 pKa = 3.29ALGIYY263 pKa = 8.9EE264 pKa = 5.05AGLAVDD270 pKa = 4.1QFNEE274 pKa = 3.89AGYY277 pKa = 11.17LNATRR282 pKa = 11.84VAIQLNEE289 pKa = 3.87NDD291 pKa = 3.63QAEE294 pKa = 4.21QLYY297 pKa = 10.5QKK299 pKa = 10.41GLKK302 pKa = 9.54NLPDD306 pKa = 3.82SQNLISNYY314 pKa = 9.6SQMLVDD320 pKa = 3.76TEE322 pKa = 4.47QYY324 pKa = 9.29MEE326 pKa = 4.64QINFLNQYY334 pKa = 9.69VSADD338 pKa = 3.89DD339 pKa = 4.57EE340 pKa = 4.89FEE342 pKa = 5.03LDD344 pKa = 3.94PKK346 pKa = 10.74EE347 pKa = 4.21YY348 pKa = 10.05WNLAQSYY355 pKa = 8.45TEE357 pKa = 3.96LEE359 pKa = 4.29RR360 pKa = 11.84YY361 pKa = 9.34EE362 pKa = 4.41MADD365 pKa = 3.44QYY367 pKa = 10.14WQAAVPFFNEE377 pKa = 3.13NSIFLKK383 pKa = 10.04EE384 pKa = 3.81AIYY387 pKa = 10.13YY388 pKa = 9.66FRR390 pKa = 11.84EE391 pKa = 4.43AGNHH395 pKa = 5.26EE396 pKa = 4.52MLSDD400 pKa = 4.31LLNKK404 pKa = 9.56YY405 pKa = 9.23VQLNPEE411 pKa = 4.54DD412 pKa = 4.94FEE414 pKa = 4.39MAQMEE419 pKa = 4.85SEE421 pKa = 4.26INDD424 pKa = 3.65LL425 pKa = 4.37
Molecular weight: 48.69 kDa
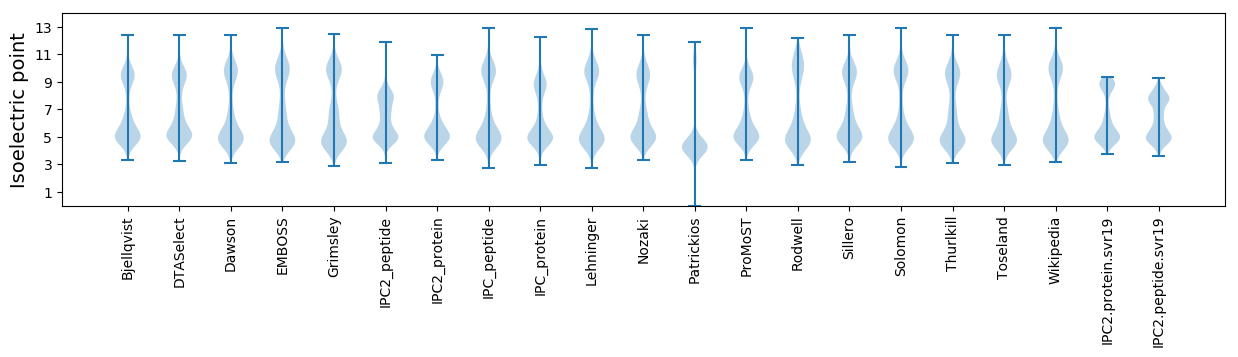
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F8I1A0|F8I1A0_WEIKK Alkaline phosphatase superfamily protein OS=Weissella koreensis (strain KACC 15510) OX=1045854 GN=WKK_03885 PE=3 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 8.34KK9 pKa = 7.47RR10 pKa = 11.84HH11 pKa = 5.71RR12 pKa = 11.84EE13 pKa = 3.7RR14 pKa = 11.84VHH16 pKa = 6.23GFRR19 pKa = 11.84KK20 pKa = 10.04RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84KK29 pKa = 8.01VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.39GRR39 pKa = 11.84KK40 pKa = 8.52VLSAA44 pKa = 4.05
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 8.34KK9 pKa = 7.47RR10 pKa = 11.84HH11 pKa = 5.71RR12 pKa = 11.84EE13 pKa = 3.7RR14 pKa = 11.84VHH16 pKa = 6.23GFRR19 pKa = 11.84KK20 pKa = 10.04RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84KK29 pKa = 8.01VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.39GRR39 pKa = 11.84KK40 pKa = 8.52VLSAA44 pKa = 4.05
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
404084 |
30 |
2007 |
300.2 |
33.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.532 ± 0.073 | 0.185 ± 0.01 |
5.89 ± 0.07 | 6.045 ± 0.065 |
4.256 ± 0.054 | 6.595 ± 0.073 |
1.918 ± 0.028 | 7.841 ± 0.079 |
6.384 ± 0.075 | 9.945 ± 0.089 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.936 ± 0.04 | 5.405 ± 0.07 |
3.38 ± 0.036 | 4.72 ± 0.068 |
3.859 ± 0.053 | 5.927 ± 0.118 |
5.728 ± 0.047 | 6.993 ± 0.055 |
1.076 ± 0.027 | 3.388 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
