
Changjiang tombus-like virus 10
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.13
Get precalculated fractions of proteins
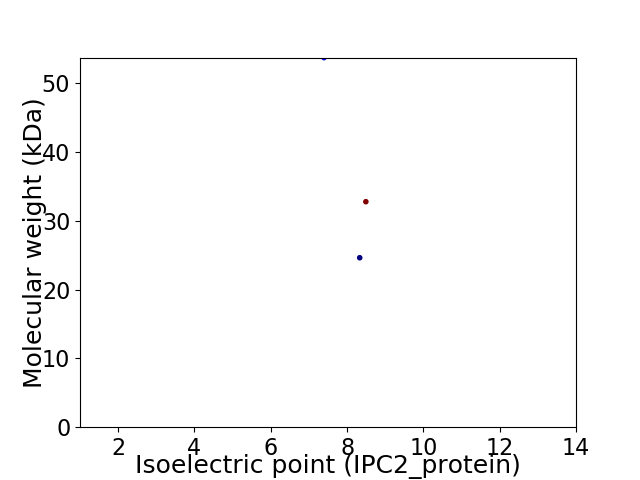
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFU0|A0A1L3KFU0_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 10 OX=1922803 PE=4 SV=1
MM1 pKa = 7.55LSGVQGPIKK10 pKa = 10.22FLIHH14 pKa = 6.36NNSATNLRR22 pKa = 11.84RR23 pKa = 11.84ALMEE27 pKa = 3.66RR28 pKa = 11.84VYY30 pKa = 10.62HH31 pKa = 6.07VEE33 pKa = 4.87ANGQLQRR40 pKa = 11.84PPQPMPGIYY49 pKa = 8.84ATLTALRR56 pKa = 11.84DD57 pKa = 4.06KK58 pKa = 10.46IAHH61 pKa = 6.06RR62 pKa = 11.84VGAATPMEE70 pKa = 3.76RR71 pKa = 11.84HH72 pKa = 5.65YY73 pKa = 10.9FVSTRR78 pKa = 11.84PADD81 pKa = 3.24KK82 pKa = 10.69RR83 pKa = 11.84RR84 pKa = 11.84IYY86 pKa = 10.39EE87 pKa = 3.61NAYY90 pKa = 10.53ASLQLTPPSVKK101 pKa = 10.13DD102 pKa = 3.27ARR104 pKa = 11.84VYY106 pKa = 10.71GAFVKK111 pKa = 10.65CEE113 pKa = 4.26KK114 pKa = 10.46INASKK119 pKa = 10.8KK120 pKa = 9.74SDD122 pKa = 3.53PAPRR126 pKa = 11.84IIQPRR131 pKa = 11.84SPRR134 pKa = 11.84YY135 pKa = 8.12NIEE138 pKa = 3.57VGRR141 pKa = 11.84YY142 pKa = 8.35LAVIEE147 pKa = 4.41HH148 pKa = 7.05DD149 pKa = 3.77VYY151 pKa = 11.58AAIDD155 pKa = 4.09EE156 pKa = 4.74LWGSTTVMKK165 pKa = 10.49GYY167 pKa = 10.12NVEE170 pKa = 4.16QIGEE174 pKa = 4.23IISAKK179 pKa = 9.21WGEE182 pKa = 4.03FTKK185 pKa = 10.63PVALGLDD192 pKa = 3.57ASRR195 pKa = 11.84FDD197 pKa = 3.23QHH199 pKa = 8.34VSVEE203 pKa = 4.09ALEE206 pKa = 4.58FEE208 pKa = 4.49HH209 pKa = 7.02GLYY212 pKa = 8.78NTIFKK217 pKa = 10.66SRR219 pKa = 11.84EE220 pKa = 3.64LRR222 pKa = 11.84RR223 pKa = 11.84LLRR226 pKa = 11.84WQLDD230 pKa = 3.66NRR232 pKa = 11.84GVAHH236 pKa = 7.39ADD238 pKa = 3.2DD239 pKa = 3.75ATFVYY244 pKa = 9.89RR245 pKa = 11.84KK246 pKa = 8.9PGSRR250 pKa = 11.84MSGDD254 pKa = 3.32MNTALGNIIIMCILVKK270 pKa = 9.79QYY272 pKa = 10.98CDD274 pKa = 3.12EE275 pKa = 4.41KK276 pKa = 11.08GIRR279 pKa = 11.84AEE281 pKa = 4.85LINNGDD287 pKa = 3.72DD288 pKa = 3.43CTLIFEE294 pKa = 4.88AEE296 pKa = 4.42HH297 pKa = 6.18VDD299 pKa = 3.27AMRR302 pKa = 11.84DD303 pKa = 3.63GLHH306 pKa = 6.07EE307 pKa = 3.92WFLRR311 pKa = 11.84YY312 pKa = 9.67GFNIVEE318 pKa = 4.11EE319 pKa = 4.8PIVDD323 pKa = 4.09VIEE326 pKa = 4.88KK327 pKa = 10.31IEE329 pKa = 4.24FCQMHH334 pKa = 6.73PVLVGDD340 pKa = 3.8SYY342 pKa = 12.27KK343 pKa = 9.95MVRR346 pKa = 11.84NFWASLSKK354 pKa = 10.67DD355 pKa = 3.32AISIRR360 pKa = 11.84SRR362 pKa = 11.84SITEE366 pKa = 3.71LQQWMHH372 pKa = 6.31CVGKK376 pKa = 10.45CGLAAASGVPIQQAYY391 pKa = 9.64YY392 pKa = 10.62SYY394 pKa = 10.88FVRR397 pKa = 11.84NGVKK401 pKa = 10.18GKK403 pKa = 10.44RR404 pKa = 11.84LDD406 pKa = 3.71QVEE409 pKa = 4.67GRR411 pKa = 11.84CGLTWFSRR419 pKa = 11.84GLHH422 pKa = 5.47ATTTHH427 pKa = 5.8ITDD430 pKa = 3.55STRR433 pKa = 11.84HH434 pKa = 4.87SFYY437 pKa = 10.88LAFGIHH443 pKa = 6.99ADD445 pKa = 3.47MQFALEE451 pKa = 4.12AEE453 pKa = 4.35YY454 pKa = 11.21DD455 pKa = 3.87RR456 pKa = 11.84LVLSTNAPKK465 pKa = 10.78AEE467 pKa = 4.33DD468 pKa = 3.91YY469 pKa = 10.7IVYY472 pKa = 10.1YY473 pKa = 10.9
MM1 pKa = 7.55LSGVQGPIKK10 pKa = 10.22FLIHH14 pKa = 6.36NNSATNLRR22 pKa = 11.84RR23 pKa = 11.84ALMEE27 pKa = 3.66RR28 pKa = 11.84VYY30 pKa = 10.62HH31 pKa = 6.07VEE33 pKa = 4.87ANGQLQRR40 pKa = 11.84PPQPMPGIYY49 pKa = 8.84ATLTALRR56 pKa = 11.84DD57 pKa = 4.06KK58 pKa = 10.46IAHH61 pKa = 6.06RR62 pKa = 11.84VGAATPMEE70 pKa = 3.76RR71 pKa = 11.84HH72 pKa = 5.65YY73 pKa = 10.9FVSTRR78 pKa = 11.84PADD81 pKa = 3.24KK82 pKa = 10.69RR83 pKa = 11.84RR84 pKa = 11.84IYY86 pKa = 10.39EE87 pKa = 3.61NAYY90 pKa = 10.53ASLQLTPPSVKK101 pKa = 10.13DD102 pKa = 3.27ARR104 pKa = 11.84VYY106 pKa = 10.71GAFVKK111 pKa = 10.65CEE113 pKa = 4.26KK114 pKa = 10.46INASKK119 pKa = 10.8KK120 pKa = 9.74SDD122 pKa = 3.53PAPRR126 pKa = 11.84IIQPRR131 pKa = 11.84SPRR134 pKa = 11.84YY135 pKa = 8.12NIEE138 pKa = 3.57VGRR141 pKa = 11.84YY142 pKa = 8.35LAVIEE147 pKa = 4.41HH148 pKa = 7.05DD149 pKa = 3.77VYY151 pKa = 11.58AAIDD155 pKa = 4.09EE156 pKa = 4.74LWGSTTVMKK165 pKa = 10.49GYY167 pKa = 10.12NVEE170 pKa = 4.16QIGEE174 pKa = 4.23IISAKK179 pKa = 9.21WGEE182 pKa = 4.03FTKK185 pKa = 10.63PVALGLDD192 pKa = 3.57ASRR195 pKa = 11.84FDD197 pKa = 3.23QHH199 pKa = 8.34VSVEE203 pKa = 4.09ALEE206 pKa = 4.58FEE208 pKa = 4.49HH209 pKa = 7.02GLYY212 pKa = 8.78NTIFKK217 pKa = 10.66SRR219 pKa = 11.84EE220 pKa = 3.64LRR222 pKa = 11.84RR223 pKa = 11.84LLRR226 pKa = 11.84WQLDD230 pKa = 3.66NRR232 pKa = 11.84GVAHH236 pKa = 7.39ADD238 pKa = 3.2DD239 pKa = 3.75ATFVYY244 pKa = 9.89RR245 pKa = 11.84KK246 pKa = 8.9PGSRR250 pKa = 11.84MSGDD254 pKa = 3.32MNTALGNIIIMCILVKK270 pKa = 9.79QYY272 pKa = 10.98CDD274 pKa = 3.12EE275 pKa = 4.41KK276 pKa = 11.08GIRR279 pKa = 11.84AEE281 pKa = 4.85LINNGDD287 pKa = 3.72DD288 pKa = 3.43CTLIFEE294 pKa = 4.88AEE296 pKa = 4.42HH297 pKa = 6.18VDD299 pKa = 3.27AMRR302 pKa = 11.84DD303 pKa = 3.63GLHH306 pKa = 6.07EE307 pKa = 3.92WFLRR311 pKa = 11.84YY312 pKa = 9.67GFNIVEE318 pKa = 4.11EE319 pKa = 4.8PIVDD323 pKa = 4.09VIEE326 pKa = 4.88KK327 pKa = 10.31IEE329 pKa = 4.24FCQMHH334 pKa = 6.73PVLVGDD340 pKa = 3.8SYY342 pKa = 12.27KK343 pKa = 9.95MVRR346 pKa = 11.84NFWASLSKK354 pKa = 10.67DD355 pKa = 3.32AISIRR360 pKa = 11.84SRR362 pKa = 11.84SITEE366 pKa = 3.71LQQWMHH372 pKa = 6.31CVGKK376 pKa = 10.45CGLAAASGVPIQQAYY391 pKa = 9.64YY392 pKa = 10.62SYY394 pKa = 10.88FVRR397 pKa = 11.84NGVKK401 pKa = 10.18GKK403 pKa = 10.44RR404 pKa = 11.84LDD406 pKa = 3.71QVEE409 pKa = 4.67GRR411 pKa = 11.84CGLTWFSRR419 pKa = 11.84GLHH422 pKa = 5.47ATTTHH427 pKa = 5.8ITDD430 pKa = 3.55STRR433 pKa = 11.84HH434 pKa = 4.87SFYY437 pKa = 10.88LAFGIHH443 pKa = 6.99ADD445 pKa = 3.47MQFALEE451 pKa = 4.12AEE453 pKa = 4.35YY454 pKa = 11.21DD455 pKa = 3.87RR456 pKa = 11.84LVLSTNAPKK465 pKa = 10.78AEE467 pKa = 4.33DD468 pKa = 3.91YY469 pKa = 10.7IVYY472 pKa = 10.1YY473 pKa = 10.9
Molecular weight: 53.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFS5|A0A1L3KFS5_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 10 OX=1922803 PE=4 SV=1
MM1 pKa = 7.63ANKK4 pKa = 8.31LTKK7 pKa = 9.44ATRR10 pKa = 11.84KK11 pKa = 9.28VKK13 pKa = 10.33RR14 pKa = 11.84AAKK17 pKa = 9.66RR18 pKa = 11.84VAKK21 pKa = 9.92AAKK24 pKa = 9.58PYY26 pKa = 10.41LPKK29 pKa = 10.74FNVKK33 pKa = 10.07KK34 pKa = 10.73GLACLDD40 pKa = 3.66PAGQKK45 pKa = 8.93AANMLLDD52 pKa = 3.79PCNAQLSPAAYY63 pKa = 10.08RR64 pKa = 11.84GDD66 pKa = 3.25QGYY69 pKa = 7.63KK70 pKa = 9.14TRR72 pKa = 11.84FVSNGSIGQAPNSTCAGIIFTPSLGRR98 pKa = 11.84VYY100 pKa = 10.79SADD103 pKa = 3.28ATTSTATVTWNVGSFPNPGQNFMQGNADD131 pKa = 4.01SIRR134 pKa = 11.84SLGACLSCYY143 pKa = 9.48PVSSVMNTSGFVYY156 pKa = 9.88TGIVPEE162 pKa = 4.36SAALAATTLDD172 pKa = 4.78GLSQLCNRR180 pKa = 11.84TSRR183 pKa = 11.84IQVAEE188 pKa = 3.88PMEE191 pKa = 4.52TKK193 pKa = 10.39FVPGAGDD200 pKa = 3.32EE201 pKa = 4.55VYY203 pKa = 10.99NPVGGSIQNDD213 pKa = 3.74NDD215 pKa = 3.65TMCIVFVFTGFSSASGIRR233 pKa = 11.84IRR235 pKa = 11.84ATNIIEE241 pKa = 4.23WKK243 pKa = 9.15PDD245 pKa = 3.45PSLGIVSEE253 pKa = 4.33SHH255 pKa = 5.66LTNPSVNTIEE265 pKa = 4.15HH266 pKa = 5.66VKK268 pKa = 10.46RR269 pKa = 11.84CLFEE273 pKa = 4.95QDD275 pKa = 3.57PHH277 pKa = 6.09WFSSVGNIVKK287 pKa = 8.97RR288 pKa = 11.84TVGGYY293 pKa = 8.3MSGGIAGAVFGGLGSLSLL311 pKa = 3.98
MM1 pKa = 7.63ANKK4 pKa = 8.31LTKK7 pKa = 9.44ATRR10 pKa = 11.84KK11 pKa = 9.28VKK13 pKa = 10.33RR14 pKa = 11.84AAKK17 pKa = 9.66RR18 pKa = 11.84VAKK21 pKa = 9.92AAKK24 pKa = 9.58PYY26 pKa = 10.41LPKK29 pKa = 10.74FNVKK33 pKa = 10.07KK34 pKa = 10.73GLACLDD40 pKa = 3.66PAGQKK45 pKa = 8.93AANMLLDD52 pKa = 3.79PCNAQLSPAAYY63 pKa = 10.08RR64 pKa = 11.84GDD66 pKa = 3.25QGYY69 pKa = 7.63KK70 pKa = 9.14TRR72 pKa = 11.84FVSNGSIGQAPNSTCAGIIFTPSLGRR98 pKa = 11.84VYY100 pKa = 10.79SADD103 pKa = 3.28ATTSTATVTWNVGSFPNPGQNFMQGNADD131 pKa = 4.01SIRR134 pKa = 11.84SLGACLSCYY143 pKa = 9.48PVSSVMNTSGFVYY156 pKa = 9.88TGIVPEE162 pKa = 4.36SAALAATTLDD172 pKa = 4.78GLSQLCNRR180 pKa = 11.84TSRR183 pKa = 11.84IQVAEE188 pKa = 3.88PMEE191 pKa = 4.52TKK193 pKa = 10.39FVPGAGDD200 pKa = 3.32EE201 pKa = 4.55VYY203 pKa = 10.99NPVGGSIQNDD213 pKa = 3.74NDD215 pKa = 3.65TMCIVFVFTGFSSASGIRR233 pKa = 11.84IRR235 pKa = 11.84ATNIIEE241 pKa = 4.23WKK243 pKa = 9.15PDD245 pKa = 3.45PSLGIVSEE253 pKa = 4.33SHH255 pKa = 5.66LTNPSVNTIEE265 pKa = 4.15HH266 pKa = 5.66VKK268 pKa = 10.46RR269 pKa = 11.84CLFEE273 pKa = 4.95QDD275 pKa = 3.57PHH277 pKa = 6.09WFSSVGNIVKK287 pKa = 8.97RR288 pKa = 11.84TVGGYY293 pKa = 8.3MSGGIAGAVFGGLGSLSLL311 pKa = 3.98
Molecular weight: 32.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1001 |
217 |
473 |
333.7 |
37.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.191 ± 0.318 | 1.798 ± 0.358 |
4.995 ± 0.645 | 5.594 ± 1.357 |
3.796 ± 0.205 | 7.692 ± 0.94 |
2.198 ± 0.722 | 5.894 ± 0.669 |
4.895 ± 0.148 | 7.992 ± 1.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.298 ± 0.334 | 4.396 ± 0.707 |
4.296 ± 0.666 | 3.397 ± 0.12 |
7.393 ± 1.672 | 6.693 ± 1.222 |
5.195 ± 0.768 | 7.592 ± 0.439 |
1.299 ± 0.14 | 3.397 ± 0.93 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
