
Dishui Lake virophage 3
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Maveriviricetes; Priklausovirales; Lavidaviridae; unclassified Lavidaviridae
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
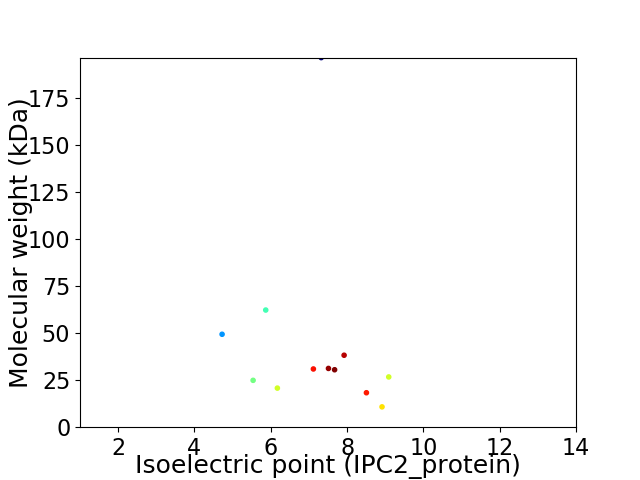
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G6XN60|A0A6G6XN60_9VIRU Putative major capsid protein OS=Dishui Lake virophage 3 OX=2704066 PE=4 SV=1
MM1 pKa = 6.42TTRR4 pKa = 11.84GNLTSADD11 pKa = 4.25PYY13 pKa = 10.97NLYY16 pKa = 10.92YY17 pKa = 10.57DD18 pKa = 3.7INVVSDD24 pKa = 3.82YY25 pKa = 11.42NPTLVGTTAPPLTFNEE41 pKa = 3.77IRR43 pKa = 11.84QNPIIKK49 pKa = 10.47YY50 pKa = 9.54PEE52 pKa = 5.12DD53 pKa = 3.49YY54 pKa = 10.51LLSVVRR60 pKa = 11.84FSIEE64 pKa = 4.22TPTLPIFIPQVLLGQPNPNKK84 pKa = 10.47LIYY87 pKa = 9.58AWGMSVTDD95 pKa = 3.8YY96 pKa = 11.18SAVGTPTYY104 pKa = 10.48YY105 pKa = 10.95LPAQEE110 pKa = 3.88NWIYY114 pKa = 10.77IPDD117 pKa = 4.94DD118 pKa = 3.48LTIPPPQGALTFQDD132 pKa = 3.75LTTEE136 pKa = 4.11YY137 pKa = 10.48YY138 pKa = 9.99YY139 pKa = 11.55VNEE142 pKa = 4.24FSLILQYY149 pKa = 11.71ANNALKK155 pKa = 10.6AAFDD159 pKa = 3.76NFNTYY164 pKa = 8.44LTGIGQPTLGAFGGVPANIAQNYY187 pKa = 7.5CPQMTYY193 pKa = 11.01DD194 pKa = 3.52PRR196 pKa = 11.84GEE198 pKa = 4.11LFSLSFPLCPPALAGAAPPFAYY220 pKa = 8.5DD221 pKa = 3.78TYY223 pKa = 11.34DD224 pKa = 3.26QNLANTAGFAGRR236 pKa = 11.84VIKK239 pKa = 10.66LYY241 pKa = 10.72MNTPLSNLLNSFPTVFQGNQQFQLTTGTEE270 pKa = 4.04DD271 pKa = 3.41MIVVYY276 pKa = 10.69NNQYY280 pKa = 10.85QNTNGGSRR288 pKa = 11.84PTYY291 pKa = 9.41PLTPVASTVAAIPQIIVPQEE311 pKa = 3.66HH312 pKa = 6.34STTILFSPISSLVFSTSLLPVQNTLLSKK340 pKa = 9.79PAIFNFYY347 pKa = 11.07DD348 pKa = 3.65GVTSSNLRR356 pKa = 11.84SSGNNNVTAPVLTDD370 pKa = 4.13FEE372 pKa = 4.72LQGATGTSSQTRR384 pKa = 11.84ITYY387 pKa = 8.39TPTAEE392 pKa = 3.92YY393 pKa = 11.03RR394 pKa = 11.84MLDD397 pKa = 3.46LRR399 pKa = 11.84GTTPVNAVEE408 pKa = 5.21VSVFWKK414 pKa = 10.67DD415 pKa = 3.22KK416 pKa = 10.61FSGLHH421 pKa = 5.86RR422 pKa = 11.84FNLAAGCAASIKK434 pKa = 10.3ILFRR438 pKa = 11.84RR439 pKa = 11.84KK440 pKa = 10.0DD441 pKa = 3.69FYY443 pKa = 11.55NATIDD448 pKa = 3.32
MM1 pKa = 6.42TTRR4 pKa = 11.84GNLTSADD11 pKa = 4.25PYY13 pKa = 10.97NLYY16 pKa = 10.92YY17 pKa = 10.57DD18 pKa = 3.7INVVSDD24 pKa = 3.82YY25 pKa = 11.42NPTLVGTTAPPLTFNEE41 pKa = 3.77IRR43 pKa = 11.84QNPIIKK49 pKa = 10.47YY50 pKa = 9.54PEE52 pKa = 5.12DD53 pKa = 3.49YY54 pKa = 10.51LLSVVRR60 pKa = 11.84FSIEE64 pKa = 4.22TPTLPIFIPQVLLGQPNPNKK84 pKa = 10.47LIYY87 pKa = 9.58AWGMSVTDD95 pKa = 3.8YY96 pKa = 11.18SAVGTPTYY104 pKa = 10.48YY105 pKa = 10.95LPAQEE110 pKa = 3.88NWIYY114 pKa = 10.77IPDD117 pKa = 4.94DD118 pKa = 3.48LTIPPPQGALTFQDD132 pKa = 3.75LTTEE136 pKa = 4.11YY137 pKa = 10.48YY138 pKa = 9.99YY139 pKa = 11.55VNEE142 pKa = 4.24FSLILQYY149 pKa = 11.71ANNALKK155 pKa = 10.6AAFDD159 pKa = 3.76NFNTYY164 pKa = 8.44LTGIGQPTLGAFGGVPANIAQNYY187 pKa = 7.5CPQMTYY193 pKa = 11.01DD194 pKa = 3.52PRR196 pKa = 11.84GEE198 pKa = 4.11LFSLSFPLCPPALAGAAPPFAYY220 pKa = 8.5DD221 pKa = 3.78TYY223 pKa = 11.34DD224 pKa = 3.26QNLANTAGFAGRR236 pKa = 11.84VIKK239 pKa = 10.66LYY241 pKa = 10.72MNTPLSNLLNSFPTVFQGNQQFQLTTGTEE270 pKa = 4.04DD271 pKa = 3.41MIVVYY276 pKa = 10.69NNQYY280 pKa = 10.85QNTNGGSRR288 pKa = 11.84PTYY291 pKa = 9.41PLTPVASTVAAIPQIIVPQEE311 pKa = 3.66HH312 pKa = 6.34STTILFSPISSLVFSTSLLPVQNTLLSKK340 pKa = 9.79PAIFNFYY347 pKa = 11.07DD348 pKa = 3.65GVTSSNLRR356 pKa = 11.84SSGNNNVTAPVLTDD370 pKa = 4.13FEE372 pKa = 4.72LQGATGTSSQTRR384 pKa = 11.84ITYY387 pKa = 8.39TPTAEE392 pKa = 3.92YY393 pKa = 11.03RR394 pKa = 11.84MLDD397 pKa = 3.46LRR399 pKa = 11.84GTTPVNAVEE408 pKa = 5.21VSVFWKK414 pKa = 10.67DD415 pKa = 3.22KK416 pKa = 10.61FSGLHH421 pKa = 5.86RR422 pKa = 11.84FNLAAGCAASIKK434 pKa = 10.3ILFRR438 pKa = 11.84RR439 pKa = 11.84KK440 pKa = 10.0DD441 pKa = 3.69FYY443 pKa = 11.55NATIDD448 pKa = 3.32
Molecular weight: 49.39 kDa
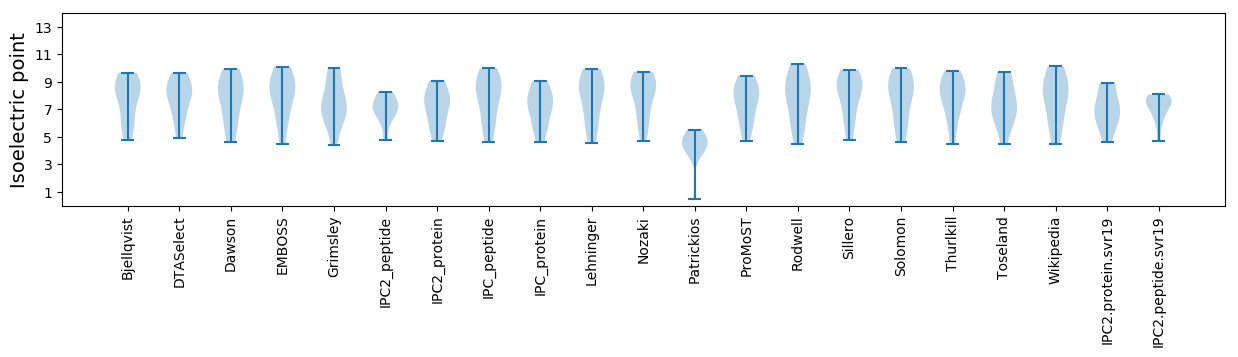
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G6XMN2|A0A6G6XMN2_9VIRU Uncharacterized protein OS=Dishui Lake virophage 3 OX=2704066 PE=4 SV=1
MM1 pKa = 7.18ITQTLINAFFRR12 pKa = 11.84VSPATTPSPPPSPDD26 pKa = 3.48PLTDD30 pKa = 3.06DD31 pKa = 3.13WVVIDD36 pKa = 3.98EE37 pKa = 4.42RR38 pKa = 11.84FGVRR42 pKa = 11.84KK43 pKa = 10.06DD44 pKa = 3.22NYY46 pKa = 9.56AINRR50 pKa = 11.84IGQVKK55 pKa = 9.59NLKK58 pKa = 9.73FNKK61 pKa = 9.33ILKK64 pKa = 10.29SYY66 pKa = 8.17FCKK69 pKa = 10.57SIGYY73 pKa = 8.32MCVSLKK79 pKa = 10.73RR80 pKa = 11.84SMDD83 pKa = 3.11ASGNLVRR90 pKa = 11.84PNNGYY95 pKa = 9.37GRR97 pKa = 11.84KK98 pKa = 8.67PVKK101 pKa = 9.47DD102 pKa = 3.56TPNNEE107 pKa = 3.77TLMLVHH113 pKa = 6.96RR114 pKa = 11.84LVGNVFIPNTNPNYY128 pKa = 10.46RR129 pKa = 11.84IIDD132 pKa = 4.31HH133 pKa = 7.24IDD135 pKa = 3.28GNKK138 pKa = 10.21LNNDD142 pKa = 3.29YY143 pKa = 11.33RR144 pKa = 11.84NLRR147 pKa = 11.84WCDD150 pKa = 3.16QRR152 pKa = 11.84RR153 pKa = 11.84NANNAKK159 pKa = 10.18SNKK162 pKa = 9.34KK163 pKa = 8.42YY164 pKa = 10.13WGVRR168 pKa = 11.84WAKK171 pKa = 10.85NMEE174 pKa = 4.04KK175 pKa = 9.73WSASIQTTVNYY186 pKa = 10.35NPNEE190 pKa = 4.29TFTHH194 pKa = 6.54FLGHH198 pKa = 6.47YY199 pKa = 9.19DD200 pKa = 4.09DD201 pKa = 5.23EE202 pKa = 4.99VMAARR207 pKa = 11.84VVKK210 pKa = 10.64KK211 pKa = 10.23FMTEE215 pKa = 3.68NYY217 pKa = 8.23PNEE220 pKa = 4.08MTGGRR225 pKa = 11.84RR226 pKa = 11.84FIEE229 pKa = 4.34EE230 pKa = 3.77
MM1 pKa = 7.18ITQTLINAFFRR12 pKa = 11.84VSPATTPSPPPSPDD26 pKa = 3.48PLTDD30 pKa = 3.06DD31 pKa = 3.13WVVIDD36 pKa = 3.98EE37 pKa = 4.42RR38 pKa = 11.84FGVRR42 pKa = 11.84KK43 pKa = 10.06DD44 pKa = 3.22NYY46 pKa = 9.56AINRR50 pKa = 11.84IGQVKK55 pKa = 9.59NLKK58 pKa = 9.73FNKK61 pKa = 9.33ILKK64 pKa = 10.29SYY66 pKa = 8.17FCKK69 pKa = 10.57SIGYY73 pKa = 8.32MCVSLKK79 pKa = 10.73RR80 pKa = 11.84SMDD83 pKa = 3.11ASGNLVRR90 pKa = 11.84PNNGYY95 pKa = 9.37GRR97 pKa = 11.84KK98 pKa = 8.67PVKK101 pKa = 9.47DD102 pKa = 3.56TPNNEE107 pKa = 3.77TLMLVHH113 pKa = 6.96RR114 pKa = 11.84LVGNVFIPNTNPNYY128 pKa = 10.46RR129 pKa = 11.84IIDD132 pKa = 4.31HH133 pKa = 7.24IDD135 pKa = 3.28GNKK138 pKa = 10.21LNNDD142 pKa = 3.29YY143 pKa = 11.33RR144 pKa = 11.84NLRR147 pKa = 11.84WCDD150 pKa = 3.16QRR152 pKa = 11.84RR153 pKa = 11.84NANNAKK159 pKa = 10.18SNKK162 pKa = 9.34KK163 pKa = 8.42YY164 pKa = 10.13WGVRR168 pKa = 11.84WAKK171 pKa = 10.85NMEE174 pKa = 4.04KK175 pKa = 9.73WSASIQTTVNYY186 pKa = 10.35NPNEE190 pKa = 4.29TFTHH194 pKa = 6.54FLGHH198 pKa = 6.47YY199 pKa = 9.19DD200 pKa = 4.09DD201 pKa = 5.23EE202 pKa = 4.99VMAARR207 pKa = 11.84VVKK210 pKa = 10.64KK211 pKa = 10.23FMTEE215 pKa = 3.68NYY217 pKa = 8.23PNEE220 pKa = 4.08MTGGRR225 pKa = 11.84RR226 pKa = 11.84FIEE229 pKa = 4.34EE230 pKa = 3.77
Molecular weight: 26.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4760 |
92 |
1726 |
396.7 |
45.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.227 ± 1.065 | 0.987 ± 0.31 |
6.282 ± 0.349 | 6.87 ± 1.37 |
4.055 ± 0.405 | 5.819 ± 0.327 |
1.492 ± 0.425 | 5.756 ± 0.6 |
7.941 ± 1.303 | 7.731 ± 0.652 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.878 ± 0.303 | 5.567 ± 0.864 |
4.958 ± 0.492 | 3.95 ± 0.483 |
5.693 ± 0.586 | 5.798 ± 0.7 |
5.945 ± 0.741 | 5.903 ± 0.298 |
1.239 ± 0.157 | 3.908 ± 0.427 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
