
Synechococcus sp. WH 5701
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Synechococcaceae; Synechococcus; unclassified Synechococcus
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
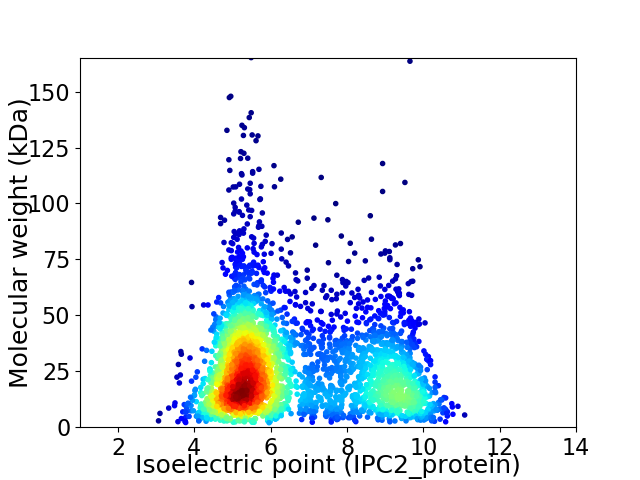
Virtual 2D-PAGE plot for 3316 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A3Z0H3|A3Z0H3_9SYNE V-type ATP synthase alpha chain OS=Synechococcus sp. WH 5701 OX=69042 GN=atpA PE=3 SV=1
MM1 pKa = 7.59ALLRR5 pKa = 11.84KK6 pKa = 9.54AFQLWDD12 pKa = 3.55EE13 pKa = 4.41ALEE16 pKa = 4.19SVTFQEE22 pKa = 4.92VSPNDD27 pKa = 3.54GGAQIQVGLGEE38 pKa = 4.48TNGLSGYY45 pKa = 9.24WSATWSTVRR54 pKa = 11.84TSASITLAPNLDD66 pKa = 3.66PALFAVAAAHH76 pKa = 6.92EE77 pKa = 4.28IANVLGLGDD86 pKa = 3.97IDD88 pKa = 3.62GGGYY92 pKa = 9.51VEE94 pKa = 5.8SLTSDD99 pKa = 3.72PFSSFHH105 pKa = 6.86LSNPNLNYY113 pKa = 10.43LSRR116 pKa = 11.84IDD118 pKa = 4.96FEE120 pKa = 6.23LINQLYY126 pKa = 10.62GEE128 pKa = 5.31DD129 pKa = 3.51STDD132 pKa = 3.41YY133 pKa = 11.09LGYY136 pKa = 10.52LAGLSIFSQVIVGDD150 pKa = 4.22SNSALAEE157 pKa = 4.34SFSGTDD163 pKa = 3.09GADD166 pKa = 4.31LIRR169 pKa = 11.84GYY171 pKa = 11.12AGDD174 pKa = 3.79DD175 pKa = 3.5TLIGGSGNDD184 pKa = 4.69LIFAGDD190 pKa = 3.73GRR192 pKa = 11.84DD193 pKa = 4.01LIIGGDD199 pKa = 3.85GADD202 pKa = 3.43VMHH205 pKa = 7.04GGFGANIFGDD215 pKa = 3.89SRR217 pKa = 11.84DD218 pKa = 3.54GSVDD222 pKa = 3.32QILIHH227 pKa = 6.84SDD229 pKa = 3.15HH230 pKa = 6.86FLMNPLTGTTTDD242 pKa = 3.51NTSGEE247 pKa = 4.28YY248 pKa = 10.56VDD250 pKa = 4.69ILEE253 pKa = 5.15GLDD256 pKa = 3.38PSDD259 pKa = 4.75NIFIEE264 pKa = 4.41AGFGRR269 pKa = 11.84SLEE272 pKa = 4.15VRR274 pKa = 11.84DD275 pKa = 3.83TTGGLGLYY283 pKa = 10.06VDD285 pKa = 5.53GIIEE289 pKa = 4.3ALYY292 pKa = 9.8TGNDD296 pKa = 3.16LSAAQLFSMTEE307 pKa = 3.48IVAAA311 pKa = 4.38
MM1 pKa = 7.59ALLRR5 pKa = 11.84KK6 pKa = 9.54AFQLWDD12 pKa = 3.55EE13 pKa = 4.41ALEE16 pKa = 4.19SVTFQEE22 pKa = 4.92VSPNDD27 pKa = 3.54GGAQIQVGLGEE38 pKa = 4.48TNGLSGYY45 pKa = 9.24WSATWSTVRR54 pKa = 11.84TSASITLAPNLDD66 pKa = 3.66PALFAVAAAHH76 pKa = 6.92EE77 pKa = 4.28IANVLGLGDD86 pKa = 3.97IDD88 pKa = 3.62GGGYY92 pKa = 9.51VEE94 pKa = 5.8SLTSDD99 pKa = 3.72PFSSFHH105 pKa = 6.86LSNPNLNYY113 pKa = 10.43LSRR116 pKa = 11.84IDD118 pKa = 4.96FEE120 pKa = 6.23LINQLYY126 pKa = 10.62GEE128 pKa = 5.31DD129 pKa = 3.51STDD132 pKa = 3.41YY133 pKa = 11.09LGYY136 pKa = 10.52LAGLSIFSQVIVGDD150 pKa = 4.22SNSALAEE157 pKa = 4.34SFSGTDD163 pKa = 3.09GADD166 pKa = 4.31LIRR169 pKa = 11.84GYY171 pKa = 11.12AGDD174 pKa = 3.79DD175 pKa = 3.5TLIGGSGNDD184 pKa = 4.69LIFAGDD190 pKa = 3.73GRR192 pKa = 11.84DD193 pKa = 4.01LIIGGDD199 pKa = 3.85GADD202 pKa = 3.43VMHH205 pKa = 7.04GGFGANIFGDD215 pKa = 3.89SRR217 pKa = 11.84DD218 pKa = 3.54GSVDD222 pKa = 3.32QILIHH227 pKa = 6.84SDD229 pKa = 3.15HH230 pKa = 6.86FLMNPLTGTTTDD242 pKa = 3.51NTSGEE247 pKa = 4.28YY248 pKa = 10.56VDD250 pKa = 4.69ILEE253 pKa = 5.15GLDD256 pKa = 3.38PSDD259 pKa = 4.75NIFIEE264 pKa = 4.41AGFGRR269 pKa = 11.84SLEE272 pKa = 4.15VRR274 pKa = 11.84DD275 pKa = 3.83TTGGLGLYY283 pKa = 10.06VDD285 pKa = 5.53GIIEE289 pKa = 4.3ALYY292 pKa = 9.8TGNDD296 pKa = 3.16LSAAQLFSMTEE307 pKa = 3.48IVAAA311 pKa = 4.38
Molecular weight: 32.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A3Z0T2|A3Z0T2_9SYNE Glutamyl/Glutaminyl-tRNA synthetase OS=Synechococcus sp. WH 5701 OX=69042 GN=WH5701_06431 PE=3 SV=1
MM1 pKa = 7.56TKK3 pKa = 9.03RR4 pKa = 11.84TLEE7 pKa = 3.92GTSRR11 pKa = 11.84KK12 pKa = 9.24RR13 pKa = 11.84KK14 pKa = 7.95RR15 pKa = 11.84VSGFRR20 pKa = 11.84VRR22 pKa = 11.84MRR24 pKa = 11.84SHH26 pKa = 6.21TGRR29 pKa = 11.84RR30 pKa = 11.84VIRR33 pKa = 11.84TRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84ARR42 pKa = 11.84LAVV45 pKa = 3.5
MM1 pKa = 7.56TKK3 pKa = 9.03RR4 pKa = 11.84TLEE7 pKa = 3.92GTSRR11 pKa = 11.84KK12 pKa = 9.24RR13 pKa = 11.84KK14 pKa = 7.95RR15 pKa = 11.84VSGFRR20 pKa = 11.84VRR22 pKa = 11.84MRR24 pKa = 11.84SHH26 pKa = 6.21TGRR29 pKa = 11.84RR30 pKa = 11.84VIRR33 pKa = 11.84TRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84ARR42 pKa = 11.84LAVV45 pKa = 3.5
Molecular weight: 5.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
883610 |
19 |
1532 |
266.5 |
28.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.478 ± 0.052 | 1.134 ± 0.017 |
4.826 ± 0.032 | 5.934 ± 0.047 |
3.045 ± 0.029 | 8.774 ± 0.045 |
2.066 ± 0.025 | 3.925 ± 0.031 |
2.126 ± 0.03 | 13.231 ± 0.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.904 ± 0.019 | 2.149 ± 0.025 |
6.21 ± 0.037 | 4.433 ± 0.031 |
7.73 ± 0.044 | 6.125 ± 0.034 |
4.426 ± 0.031 | 6.948 ± 0.04 |
1.757 ± 0.024 | 1.779 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
