
Tropicibacter naphthalenivorans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Tropicibacter
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
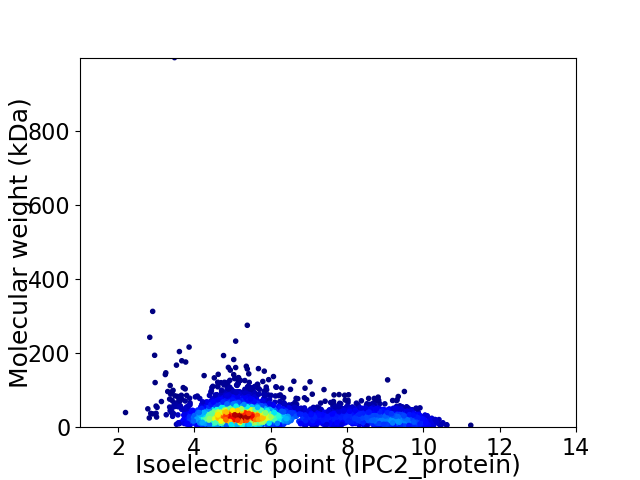
Virtual 2D-PAGE plot for 4206 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0N7M0G0|A0A0N7M0G0_9RHOB Uncharacterized protein OS=Tropicibacter naphthalenivorans OX=441103 GN=TRN7648_02949 PE=4 SV=1
MM1 pKa = 7.34PAIYY5 pKa = 8.7TWQHH9 pKa = 4.91VPTHH13 pKa = 6.64KK14 pKa = 10.54YY15 pKa = 9.36VPAYY19 pKa = 9.36DD20 pKa = 2.83QWYY23 pKa = 8.43YY24 pKa = 11.63DD25 pKa = 4.41KK26 pKa = 11.18IHH28 pKa = 6.15ITGPGAIAYY37 pKa = 8.78GGYY40 pKa = 10.58GDD42 pKa = 5.19DD43 pKa = 3.33VYY45 pKa = 11.69YY46 pKa = 10.6HH47 pKa = 6.12YY48 pKa = 11.05GVRR51 pKa = 11.84SYY53 pKa = 11.41SSPRR57 pKa = 11.84DD58 pKa = 3.42KK59 pKa = 11.24VIVEE63 pKa = 4.59FADD66 pKa = 3.07QGYY69 pKa = 10.03DD70 pKa = 3.08IVHH73 pKa = 6.6VDD75 pKa = 3.71PLADD79 pKa = 3.59IVAQLYY85 pKa = 9.17FVEE88 pKa = 5.66EE89 pKa = 3.86IHH91 pKa = 6.68LGCSSLSRR99 pKa = 11.84EE100 pKa = 5.27IYY102 pKa = 8.93GTWQAQAFYY111 pKa = 10.31GLHH114 pKa = 6.59RR115 pKa = 11.84GDD117 pKa = 3.25TLYY120 pKa = 10.94AGEE123 pKa = 5.15GDD125 pKa = 3.52DD126 pKa = 3.79TLYY129 pKa = 10.96TRR131 pKa = 11.84VDD133 pKa = 2.79VDD135 pKa = 3.65LRR137 pKa = 11.84KK138 pKa = 10.2EE139 pKa = 3.9NAEE142 pKa = 4.02FEE144 pKa = 4.73HH145 pKa = 7.72VIATYY150 pKa = 10.58DD151 pKa = 3.75GNSEE155 pKa = 3.87NSNLNFQNLGWRR167 pKa = 11.84FVRR170 pKa = 11.84GEE172 pKa = 3.84PWTVHH177 pKa = 4.34GSEE180 pKa = 4.47RR181 pKa = 11.84GDD183 pKa = 4.14DD184 pKa = 3.52IVGGGSLDD192 pKa = 3.22KK193 pKa = 10.99LYY195 pKa = 10.86GYY197 pKa = 10.7GGDD200 pKa = 4.21DD201 pKa = 3.6SLVGAGLNDD210 pKa = 3.7YY211 pKa = 11.16LSGGEE216 pKa = 4.67GNDD219 pKa = 3.42SLYY222 pKa = 11.38GGVDD226 pKa = 3.11NDD228 pKa = 4.19TLYY231 pKa = 11.64GDD233 pKa = 4.83AGDD236 pKa = 5.5DD237 pKa = 3.82YY238 pKa = 11.34LEE240 pKa = 4.69SGAGHH245 pKa = 6.97DD246 pKa = 3.87EE247 pKa = 4.63VYY249 pKa = 10.9GGTGDD254 pKa = 3.65DD255 pKa = 3.9TLIGSDD261 pKa = 4.8GNDD264 pKa = 3.16TLAGGDD270 pKa = 3.46GDD272 pKa = 4.81DD273 pKa = 4.57FYY275 pKa = 11.86RR276 pKa = 11.84DD277 pKa = 3.27VSLGSVIIEE286 pKa = 4.38DD287 pKa = 4.02ALGGDD292 pKa = 4.0DD293 pKa = 4.13TQIITQGATRR303 pKa = 11.84QGEE306 pKa = 4.23NVEE309 pKa = 3.82RR310 pKa = 11.84LIYY313 pKa = 9.65EE314 pKa = 4.18RR315 pKa = 11.84SYY317 pKa = 10.93VLGVEE322 pKa = 4.47VRR324 pKa = 11.84QDD326 pKa = 3.42LYY328 pKa = 11.88VEE330 pKa = 4.67GNSVGNSLKK339 pKa = 10.2TGIGDD344 pKa = 3.42DD345 pKa = 4.1TLRR348 pKa = 11.84GMAGDD353 pKa = 3.97DD354 pKa = 3.42TLMGGSGADD363 pKa = 3.7LYY365 pKa = 11.21EE366 pKa = 5.17GGAGRR371 pKa = 11.84DD372 pKa = 4.04FIEE375 pKa = 4.82DD376 pKa = 3.12RR377 pKa = 11.84GTRR380 pKa = 11.84TVTDD384 pKa = 4.43ADD386 pKa = 3.08TGMQVASGLDD396 pKa = 3.41DD397 pKa = 3.81TLDD400 pKa = 3.93GGLGADD406 pKa = 4.65FMRR409 pKa = 11.84GGLGSDD415 pKa = 4.4LYY417 pKa = 10.77IVDD420 pKa = 3.68NAGDD424 pKa = 3.65VALDD428 pKa = 3.57SAEE431 pKa = 4.15DD432 pKa = 3.42AGAEE436 pKa = 4.02DD437 pKa = 4.71TVWFHH442 pKa = 7.28ASEE445 pKa = 4.89GDD447 pKa = 3.34LTYY450 pKa = 10.9NASVMNMRR458 pKa = 11.84EE459 pKa = 4.09VEE461 pKa = 4.17TLRR464 pKa = 11.84IDD466 pKa = 3.89LSAAGQVSLSTDD478 pKa = 2.55ARR480 pKa = 11.84ADD482 pKa = 3.87HH483 pKa = 7.04IIGNAASDD491 pKa = 4.02TIDD494 pKa = 3.38GGFGNDD500 pKa = 3.38LNLGGRR506 pKa = 11.84GTLGRR511 pKa = 11.84DD512 pKa = 3.53GQSLSDD518 pKa = 3.6VLGEE522 pKa = 4.19EE523 pKa = 4.5VAGDD527 pKa = 3.66DD528 pKa = 5.05ARR530 pKa = 11.84THH532 pKa = 6.92DD533 pKa = 4.17VLKK536 pKa = 11.02GGAGDD541 pKa = 3.74DD542 pKa = 4.07TIFASVGDD550 pKa = 4.38SVDD553 pKa = 3.26GGAGQDD559 pKa = 3.42EE560 pKa = 5.29LFVNAAVGPDD570 pKa = 3.16QITNVEE576 pKa = 3.9RR577 pKa = 11.84TVFLEE582 pKa = 4.0ADD584 pKa = 3.5TEE586 pKa = 4.5LFVGGTGTTVITQNDD601 pKa = 3.91SFVQGNAFALTGQGVQLLAQGGDD624 pKa = 3.59DD625 pKa = 3.69SVTGTGSVFVDD636 pKa = 4.08LGTGNDD642 pKa = 3.67SDD644 pKa = 3.95THH646 pKa = 7.47LLGTGGTTRR655 pKa = 11.84TVIVGGEE662 pKa = 4.16GGDD665 pKa = 3.8TVTLPVDD672 pKa = 4.38YY673 pKa = 10.13EE674 pKa = 4.44VLWITEE680 pKa = 3.96ATATEE685 pKa = 4.68TVTRR689 pKa = 11.84WDD691 pKa = 4.0ADD693 pKa = 3.69SQSWIVAEE701 pKa = 3.9YY702 pKa = 10.46DD703 pKa = 3.74LPVLQMVGQAEE714 pKa = 4.36VAPGQIVDD722 pKa = 3.54YY723 pKa = 9.1TFAVHH728 pKa = 6.21GVEE731 pKa = 5.08HH732 pKa = 7.18IEE734 pKa = 4.0LNYY737 pKa = 10.62QGVQVSATPVEE748 pKa = 4.45LLPVAPEE755 pKa = 3.94EE756 pKa = 3.96LTPDD760 pKa = 3.48SFTATHH766 pKa = 5.91ATIEE770 pKa = 4.13AALLGDD776 pKa = 4.35PAALGYY782 pKa = 8.4GTGAQLVRR790 pKa = 11.84YY791 pKa = 8.41VISLDD796 pKa = 3.44TSVGVPVFSVMTTDD810 pKa = 3.34NNVPFDD816 pKa = 4.27GEE818 pKa = 3.81PMYY821 pKa = 10.88RR822 pKa = 11.84FVDD825 pKa = 4.04APDD828 pKa = 3.83GTPALAGLTNLSAAEE843 pKa = 3.91ATALFSFVTDD853 pKa = 3.38GDD855 pKa = 3.93GTTRR859 pKa = 11.84PGTLTLNSLPEE870 pKa = 4.61AGSDD874 pKa = 4.68LIQDD878 pKa = 4.93LSTTDD883 pKa = 3.56NPEE886 pKa = 4.26DD887 pKa = 3.87PPASVGVIWVEE898 pKa = 3.97AYY900 pKa = 10.32DD901 pKa = 3.31ITGRR905 pKa = 11.84STGAYY910 pKa = 7.72PLEE913 pKa = 4.42VNFAQIPGQWIAHH926 pKa = 5.64TSDD929 pKa = 4.66DD930 pKa = 4.07RR931 pKa = 11.84VTDD934 pKa = 4.36LSQSPASRR942 pKa = 11.84DD943 pKa = 3.75TIIGGVWNDD952 pKa = 4.13TILSGDD958 pKa = 4.04GGDD961 pKa = 3.91EE962 pKa = 4.13IYY964 pKa = 11.07GGGSDD969 pKa = 4.4HH970 pKa = 7.4SGTEE974 pKa = 3.94QIDD977 pKa = 3.72AGWGNDD983 pKa = 3.28FVQVISNGLTYY994 pKa = 10.65HH995 pKa = 6.76FDD997 pKa = 3.14INYY1000 pKa = 9.96EE1001 pKa = 3.98IVPWDD1006 pKa = 3.9AGSTPTT1012 pKa = 5.02
MM1 pKa = 7.34PAIYY5 pKa = 8.7TWQHH9 pKa = 4.91VPTHH13 pKa = 6.64KK14 pKa = 10.54YY15 pKa = 9.36VPAYY19 pKa = 9.36DD20 pKa = 2.83QWYY23 pKa = 8.43YY24 pKa = 11.63DD25 pKa = 4.41KK26 pKa = 11.18IHH28 pKa = 6.15ITGPGAIAYY37 pKa = 8.78GGYY40 pKa = 10.58GDD42 pKa = 5.19DD43 pKa = 3.33VYY45 pKa = 11.69YY46 pKa = 10.6HH47 pKa = 6.12YY48 pKa = 11.05GVRR51 pKa = 11.84SYY53 pKa = 11.41SSPRR57 pKa = 11.84DD58 pKa = 3.42KK59 pKa = 11.24VIVEE63 pKa = 4.59FADD66 pKa = 3.07QGYY69 pKa = 10.03DD70 pKa = 3.08IVHH73 pKa = 6.6VDD75 pKa = 3.71PLADD79 pKa = 3.59IVAQLYY85 pKa = 9.17FVEE88 pKa = 5.66EE89 pKa = 3.86IHH91 pKa = 6.68LGCSSLSRR99 pKa = 11.84EE100 pKa = 5.27IYY102 pKa = 8.93GTWQAQAFYY111 pKa = 10.31GLHH114 pKa = 6.59RR115 pKa = 11.84GDD117 pKa = 3.25TLYY120 pKa = 10.94AGEE123 pKa = 5.15GDD125 pKa = 3.52DD126 pKa = 3.79TLYY129 pKa = 10.96TRR131 pKa = 11.84VDD133 pKa = 2.79VDD135 pKa = 3.65LRR137 pKa = 11.84KK138 pKa = 10.2EE139 pKa = 3.9NAEE142 pKa = 4.02FEE144 pKa = 4.73HH145 pKa = 7.72VIATYY150 pKa = 10.58DD151 pKa = 3.75GNSEE155 pKa = 3.87NSNLNFQNLGWRR167 pKa = 11.84FVRR170 pKa = 11.84GEE172 pKa = 3.84PWTVHH177 pKa = 4.34GSEE180 pKa = 4.47RR181 pKa = 11.84GDD183 pKa = 4.14DD184 pKa = 3.52IVGGGSLDD192 pKa = 3.22KK193 pKa = 10.99LYY195 pKa = 10.86GYY197 pKa = 10.7GGDD200 pKa = 4.21DD201 pKa = 3.6SLVGAGLNDD210 pKa = 3.7YY211 pKa = 11.16LSGGEE216 pKa = 4.67GNDD219 pKa = 3.42SLYY222 pKa = 11.38GGVDD226 pKa = 3.11NDD228 pKa = 4.19TLYY231 pKa = 11.64GDD233 pKa = 4.83AGDD236 pKa = 5.5DD237 pKa = 3.82YY238 pKa = 11.34LEE240 pKa = 4.69SGAGHH245 pKa = 6.97DD246 pKa = 3.87EE247 pKa = 4.63VYY249 pKa = 10.9GGTGDD254 pKa = 3.65DD255 pKa = 3.9TLIGSDD261 pKa = 4.8GNDD264 pKa = 3.16TLAGGDD270 pKa = 3.46GDD272 pKa = 4.81DD273 pKa = 4.57FYY275 pKa = 11.86RR276 pKa = 11.84DD277 pKa = 3.27VSLGSVIIEE286 pKa = 4.38DD287 pKa = 4.02ALGGDD292 pKa = 4.0DD293 pKa = 4.13TQIITQGATRR303 pKa = 11.84QGEE306 pKa = 4.23NVEE309 pKa = 3.82RR310 pKa = 11.84LIYY313 pKa = 9.65EE314 pKa = 4.18RR315 pKa = 11.84SYY317 pKa = 10.93VLGVEE322 pKa = 4.47VRR324 pKa = 11.84QDD326 pKa = 3.42LYY328 pKa = 11.88VEE330 pKa = 4.67GNSVGNSLKK339 pKa = 10.2TGIGDD344 pKa = 3.42DD345 pKa = 4.1TLRR348 pKa = 11.84GMAGDD353 pKa = 3.97DD354 pKa = 3.42TLMGGSGADD363 pKa = 3.7LYY365 pKa = 11.21EE366 pKa = 5.17GGAGRR371 pKa = 11.84DD372 pKa = 4.04FIEE375 pKa = 4.82DD376 pKa = 3.12RR377 pKa = 11.84GTRR380 pKa = 11.84TVTDD384 pKa = 4.43ADD386 pKa = 3.08TGMQVASGLDD396 pKa = 3.41DD397 pKa = 3.81TLDD400 pKa = 3.93GGLGADD406 pKa = 4.65FMRR409 pKa = 11.84GGLGSDD415 pKa = 4.4LYY417 pKa = 10.77IVDD420 pKa = 3.68NAGDD424 pKa = 3.65VALDD428 pKa = 3.57SAEE431 pKa = 4.15DD432 pKa = 3.42AGAEE436 pKa = 4.02DD437 pKa = 4.71TVWFHH442 pKa = 7.28ASEE445 pKa = 4.89GDD447 pKa = 3.34LTYY450 pKa = 10.9NASVMNMRR458 pKa = 11.84EE459 pKa = 4.09VEE461 pKa = 4.17TLRR464 pKa = 11.84IDD466 pKa = 3.89LSAAGQVSLSTDD478 pKa = 2.55ARR480 pKa = 11.84ADD482 pKa = 3.87HH483 pKa = 7.04IIGNAASDD491 pKa = 4.02TIDD494 pKa = 3.38GGFGNDD500 pKa = 3.38LNLGGRR506 pKa = 11.84GTLGRR511 pKa = 11.84DD512 pKa = 3.53GQSLSDD518 pKa = 3.6VLGEE522 pKa = 4.19EE523 pKa = 4.5VAGDD527 pKa = 3.66DD528 pKa = 5.05ARR530 pKa = 11.84THH532 pKa = 6.92DD533 pKa = 4.17VLKK536 pKa = 11.02GGAGDD541 pKa = 3.74DD542 pKa = 4.07TIFASVGDD550 pKa = 4.38SVDD553 pKa = 3.26GGAGQDD559 pKa = 3.42EE560 pKa = 5.29LFVNAAVGPDD570 pKa = 3.16QITNVEE576 pKa = 3.9RR577 pKa = 11.84TVFLEE582 pKa = 4.0ADD584 pKa = 3.5TEE586 pKa = 4.5LFVGGTGTTVITQNDD601 pKa = 3.91SFVQGNAFALTGQGVQLLAQGGDD624 pKa = 3.59DD625 pKa = 3.69SVTGTGSVFVDD636 pKa = 4.08LGTGNDD642 pKa = 3.67SDD644 pKa = 3.95THH646 pKa = 7.47LLGTGGTTRR655 pKa = 11.84TVIVGGEE662 pKa = 4.16GGDD665 pKa = 3.8TVTLPVDD672 pKa = 4.38YY673 pKa = 10.13EE674 pKa = 4.44VLWITEE680 pKa = 3.96ATATEE685 pKa = 4.68TVTRR689 pKa = 11.84WDD691 pKa = 4.0ADD693 pKa = 3.69SQSWIVAEE701 pKa = 3.9YY702 pKa = 10.46DD703 pKa = 3.74LPVLQMVGQAEE714 pKa = 4.36VAPGQIVDD722 pKa = 3.54YY723 pKa = 9.1TFAVHH728 pKa = 6.21GVEE731 pKa = 5.08HH732 pKa = 7.18IEE734 pKa = 4.0LNYY737 pKa = 10.62QGVQVSATPVEE748 pKa = 4.45LLPVAPEE755 pKa = 3.94EE756 pKa = 3.96LTPDD760 pKa = 3.48SFTATHH766 pKa = 5.91ATIEE770 pKa = 4.13AALLGDD776 pKa = 4.35PAALGYY782 pKa = 8.4GTGAQLVRR790 pKa = 11.84YY791 pKa = 8.41VISLDD796 pKa = 3.44TSVGVPVFSVMTTDD810 pKa = 3.34NNVPFDD816 pKa = 4.27GEE818 pKa = 3.81PMYY821 pKa = 10.88RR822 pKa = 11.84FVDD825 pKa = 4.04APDD828 pKa = 3.83GTPALAGLTNLSAAEE843 pKa = 3.91ATALFSFVTDD853 pKa = 3.38GDD855 pKa = 3.93GTTRR859 pKa = 11.84PGTLTLNSLPEE870 pKa = 4.61AGSDD874 pKa = 4.68LIQDD878 pKa = 4.93LSTTDD883 pKa = 3.56NPEE886 pKa = 4.26DD887 pKa = 3.87PPASVGVIWVEE898 pKa = 3.97AYY900 pKa = 10.32DD901 pKa = 3.31ITGRR905 pKa = 11.84STGAYY910 pKa = 7.72PLEE913 pKa = 4.42VNFAQIPGQWIAHH926 pKa = 5.64TSDD929 pKa = 4.66DD930 pKa = 4.07RR931 pKa = 11.84VTDD934 pKa = 4.36LSQSPASRR942 pKa = 11.84DD943 pKa = 3.75TIIGGVWNDD952 pKa = 4.13TILSGDD958 pKa = 4.04GGDD961 pKa = 3.91EE962 pKa = 4.13IYY964 pKa = 11.07GGGSDD969 pKa = 4.4HH970 pKa = 7.4SGTEE974 pKa = 3.94QIDD977 pKa = 3.72AGWGNDD983 pKa = 3.28FVQVISNGLTYY994 pKa = 10.65HH995 pKa = 6.76FDD997 pKa = 3.14INYY1000 pKa = 9.96EE1001 pKa = 3.98IVPWDD1006 pKa = 3.9AGSTPTT1012 pKa = 5.02
Molecular weight: 106.74 kDa
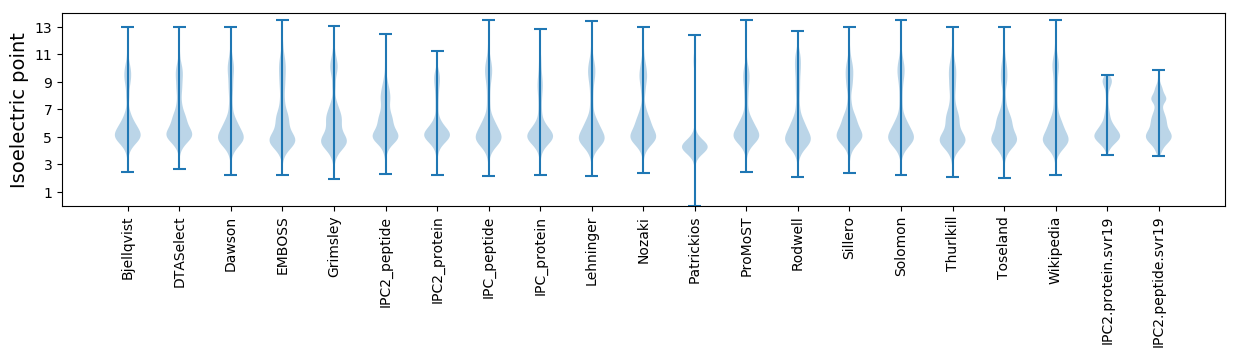
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P1GTW0|A0A0P1GTW0_9RHOB Hydroxyacylglutathione hydrolase OS=Tropicibacter naphthalenivorans OX=441103 GN=TRN7648_01509 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1340933 |
29 |
9577 |
318.8 |
34.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.321 ± 0.057 | 0.884 ± 0.012 |
6.478 ± 0.07 | 5.779 ± 0.037 |
3.659 ± 0.023 | 8.883 ± 0.066 |
2.028 ± 0.022 | 4.977 ± 0.025 |
3.197 ± 0.034 | 9.951 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.787 ± 0.024 | 2.627 ± 0.021 |
5.038 ± 0.036 | 3.455 ± 0.024 |
6.366 ± 0.051 | 4.984 ± 0.025 |
5.757 ± 0.042 | 7.204 ± 0.029 |
1.389 ± 0.016 | 2.233 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
