
Escherichia phage haarsle
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Drexlerviridae; Tempevirinae; Hanrivervirus; Escherichia virus haarsle
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
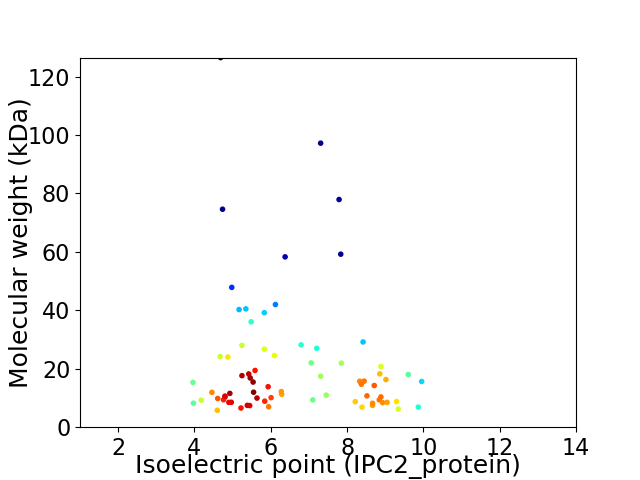
Virtual 2D-PAGE plot for 74 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6B9WV00|A0A6B9WV00_9CAUD Uncharacterized protein OS=Escherichia phage haarsle OX=2696402 GN=haarsle_1 PE=4 SV=1
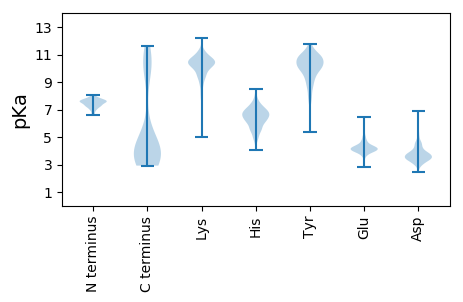
MM1 pKa = 7.0SQDD4 pKa = 3.09GFFEE8 pKa = 4.19RR9 pKa = 11.84LQMAEE14 pKa = 3.8DD15 pKa = 3.52AGLNKK20 pKa = 9.59EE21 pKa = 4.11AALEE25 pKa = 3.94VAYY28 pKa = 9.67KK29 pKa = 10.29IRR31 pKa = 11.84TLDD34 pKa = 3.72DD35 pKa = 3.96ALGEE39 pKa = 3.83MDD41 pKa = 4.5MDD43 pKa = 4.56HH44 pKa = 6.84EE45 pKa = 5.04CGAAFADD52 pKa = 3.6PTMIVNDD59 pKa = 4.6CGCNFDD65 pKa = 4.9PNCPRR70 pKa = 11.84CFPFF74 pKa = 5.73
MM1 pKa = 7.0SQDD4 pKa = 3.09GFFEE8 pKa = 4.19RR9 pKa = 11.84LQMAEE14 pKa = 3.8DD15 pKa = 3.52AGLNKK20 pKa = 9.59EE21 pKa = 4.11AALEE25 pKa = 3.94VAYY28 pKa = 9.67KK29 pKa = 10.29IRR31 pKa = 11.84TLDD34 pKa = 3.72DD35 pKa = 3.96ALGEE39 pKa = 3.83MDD41 pKa = 4.5MDD43 pKa = 4.56HH44 pKa = 6.84EE45 pKa = 5.04CGAAFADD52 pKa = 3.6PTMIVNDD59 pKa = 4.6CGCNFDD65 pKa = 4.9PNCPRR70 pKa = 11.84CFPFF74 pKa = 5.73
Molecular weight: 8.21 kDa
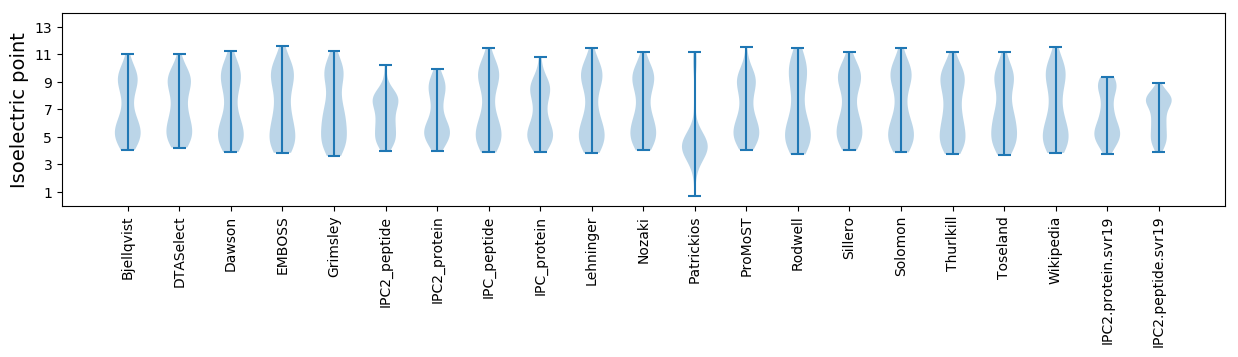
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6B9WY38|A0A6B9WY38_9CAUD Terminase small subunit OS=Escherichia phage haarsle OX=2696402 GN=haarsle_16 PE=4 SV=1
MM1 pKa = 7.72VEE3 pKa = 4.16LNDD6 pKa = 3.4IRR8 pKa = 11.84VGTKK12 pKa = 9.98FRR14 pKa = 11.84VTWADD19 pKa = 3.79EE20 pKa = 4.17YY21 pKa = 11.34CGVSKK26 pKa = 10.82GQVVTVDD33 pKa = 3.67SIYY36 pKa = 10.65RR37 pKa = 11.84GCSKK41 pKa = 10.62DD42 pKa = 3.29FRR44 pKa = 11.84RR45 pKa = 11.84PRR47 pKa = 11.84IKK49 pKa = 10.15NGYY52 pKa = 8.77IITRR56 pKa = 11.84RR57 pKa = 11.84FGFEE61 pKa = 3.49HH62 pKa = 6.91YY63 pKa = 10.56YY64 pKa = 10.73VVATQGVLIEE74 pKa = 4.1LQRR77 pKa = 11.84ISDD80 pKa = 3.56HH81 pKa = 6.57RR82 pKa = 11.84GCHH85 pKa = 4.96VKK87 pKa = 7.63TTKK90 pKa = 10.24IPFQRR95 pKa = 11.84ARR97 pKa = 11.84YY98 pKa = 7.96DD99 pKa = 3.15ARR101 pKa = 11.84RR102 pKa = 11.84MRR104 pKa = 11.84RR105 pKa = 11.84LARR108 pKa = 11.84NAIKK112 pKa = 10.27FKK114 pKa = 10.34KK115 pKa = 9.86HH116 pKa = 5.07GGNFYY121 pKa = 11.16RR122 pKa = 11.84MYY124 pKa = 10.86KK125 pKa = 10.38GIARR129 pKa = 11.84NAGKK133 pKa = 10.28
MM1 pKa = 7.72VEE3 pKa = 4.16LNDD6 pKa = 3.4IRR8 pKa = 11.84VGTKK12 pKa = 9.98FRR14 pKa = 11.84VTWADD19 pKa = 3.79EE20 pKa = 4.17YY21 pKa = 11.34CGVSKK26 pKa = 10.82GQVVTVDD33 pKa = 3.67SIYY36 pKa = 10.65RR37 pKa = 11.84GCSKK41 pKa = 10.62DD42 pKa = 3.29FRR44 pKa = 11.84RR45 pKa = 11.84PRR47 pKa = 11.84IKK49 pKa = 10.15NGYY52 pKa = 8.77IITRR56 pKa = 11.84RR57 pKa = 11.84FGFEE61 pKa = 3.49HH62 pKa = 6.91YY63 pKa = 10.56YY64 pKa = 10.73VVATQGVLIEE74 pKa = 4.1LQRR77 pKa = 11.84ISDD80 pKa = 3.56HH81 pKa = 6.57RR82 pKa = 11.84GCHH85 pKa = 4.96VKK87 pKa = 7.63TTKK90 pKa = 10.24IPFQRR95 pKa = 11.84ARR97 pKa = 11.84YY98 pKa = 7.96DD99 pKa = 3.15ARR101 pKa = 11.84RR102 pKa = 11.84MRR104 pKa = 11.84RR105 pKa = 11.84LARR108 pKa = 11.84NAIKK112 pKa = 10.27FKK114 pKa = 10.34KK115 pKa = 9.86HH116 pKa = 5.07GGNFYY121 pKa = 11.16RR122 pKa = 11.84MYY124 pKa = 10.86KK125 pKa = 10.38GIARR129 pKa = 11.84NAGKK133 pKa = 10.28
Molecular weight: 15.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14254 |
50 |
1141 |
192.6 |
21.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.18 ± 0.427 | 1.347 ± 0.172 |
6.132 ± 0.286 | 6.875 ± 0.248 |
3.887 ± 0.16 | 7.633 ± 0.307 |
1.782 ± 0.216 | 6.581 ± 0.199 |
7.415 ± 0.34 | 6.503 ± 0.197 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.136 ± 0.219 | 4.771 ± 0.226 |
3.396 ± 0.198 | 3.957 ± 0.279 |
4.918 ± 0.238 | 6.111 ± 0.242 |
5.283 ± 0.243 | 6.875 ± 0.222 |
1.459 ± 0.118 | 3.76 ± 0.17 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
