
Human cytomegalovirus (strain Merlin) (HHV-5) (Human herpesvirus 5)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Betaherpesvirinae; Cytomegalovirus; Human betaherpesvirus 5
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
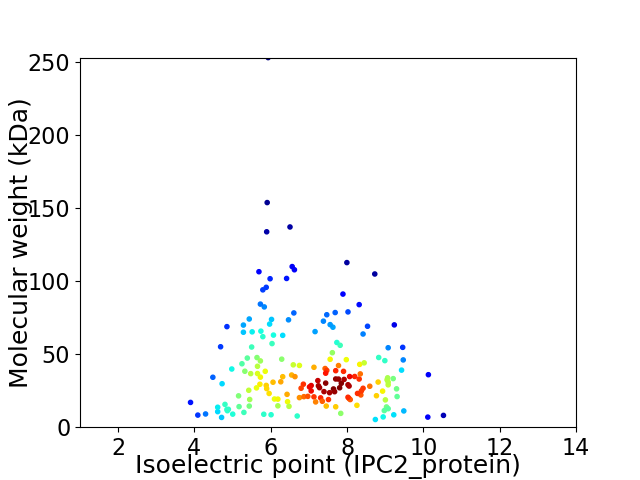
Virtual 2D-PAGE plot for 180 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
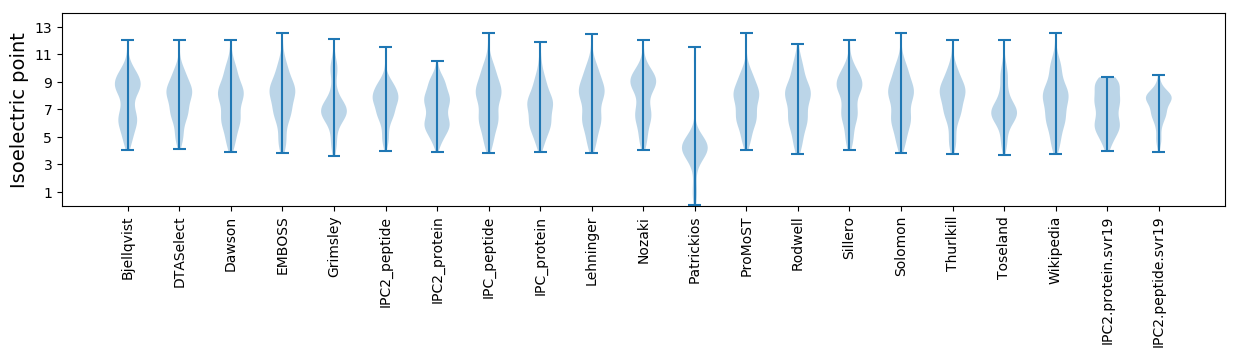
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|F5H8R6|UL96_HCMVM Protein UL96 OS=Human cytomegalovirus (strain Merlin) OX=295027 GN=UL96 PE=3 SV=1
MM1 pKa = 7.29SLFYY5 pKa = 10.51RR6 pKa = 11.84AVALGTLSALVWYY19 pKa = 7.5STSILAEE26 pKa = 4.05INEE29 pKa = 4.36NSCSSSSVDD38 pKa = 4.59HH39 pKa = 7.01EE40 pKa = 4.54DD41 pKa = 3.96CEE43 pKa = 4.58EE44 pKa = 4.11PDD46 pKa = 3.55EE47 pKa = 4.67IVRR50 pKa = 11.84EE51 pKa = 4.12EE52 pKa = 3.71QDD54 pKa = 3.41YY55 pKa = 10.84RR56 pKa = 11.84ALLAFSLVICGTLLVTCVII75 pKa = 4.52
MM1 pKa = 7.29SLFYY5 pKa = 10.51RR6 pKa = 11.84AVALGTLSALVWYY19 pKa = 7.5STSILAEE26 pKa = 4.05INEE29 pKa = 4.36NSCSSSSVDD38 pKa = 4.59HH39 pKa = 7.01EE40 pKa = 4.54DD41 pKa = 3.96CEE43 pKa = 4.58EE44 pKa = 4.11PDD46 pKa = 3.55EE47 pKa = 4.67IVRR50 pKa = 11.84EE51 pKa = 4.12EE52 pKa = 3.71QDD54 pKa = 3.41YY55 pKa = 10.84RR56 pKa = 11.84ALLAFSLVICGTLLVTCVII75 pKa = 4.52
Molecular weight: 8.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|F5HBC6|PP71_HCMVM Protein pp71 OS=Human cytomegalovirus (strain Merlin) OX=295027 GN=UL82 PE=3 SV=1
MM1 pKa = 7.61GNPRR5 pKa = 11.84SPLDD9 pKa = 3.11RR10 pKa = 11.84RR11 pKa = 11.84LRR13 pKa = 11.84FRR15 pKa = 11.84NVPFLPPRR23 pKa = 11.84IAASSTTTARR33 pKa = 11.84SLKK36 pKa = 10.05AKK38 pKa = 8.48TMEE41 pKa = 3.93MRR43 pKa = 11.84FTIAWMWFPSVLLILGLLTPPSNGCTVDD71 pKa = 2.93VGRR74 pKa = 11.84NVSIRR79 pKa = 11.84EE80 pKa = 3.74QCRR83 pKa = 11.84LRR85 pKa = 11.84NGATFSKK92 pKa = 10.98GDD94 pKa = 3.37IEE96 pKa = 5.57GNFSGPVVVEE106 pKa = 4.44LDD108 pKa = 3.82YY109 pKa = 11.77EE110 pKa = 4.91DD111 pKa = 4.89IDD113 pKa = 3.73ITGEE117 pKa = 3.9RR118 pKa = 11.84QRR120 pKa = 11.84LRR122 pKa = 11.84FHH124 pKa = 7.13LSGLGCPTKK133 pKa = 10.92EE134 pKa = 4.84NIRR137 pKa = 11.84KK138 pKa = 10.05DD139 pKa = 3.53NEE141 pKa = 4.06SDD143 pKa = 3.66VNGGIRR149 pKa = 11.84WALYY153 pKa = 9.78IQTGDD158 pKa = 2.97AKK160 pKa = 10.84YY161 pKa = 10.53GIRR164 pKa = 11.84NQHH167 pKa = 5.28LSIRR171 pKa = 11.84LMYY174 pKa = 10.14PGEE177 pKa = 4.51KK178 pKa = 8.85NTQQLLGSDD187 pKa = 4.87FSCEE191 pKa = 3.4RR192 pKa = 11.84HH193 pKa = 5.67RR194 pKa = 11.84RR195 pKa = 11.84PSTPLGKK202 pKa = 9.93NAEE205 pKa = 4.32VPPATRR211 pKa = 11.84TSSTYY216 pKa = 10.31GVLSAFVVWIGSGLNIIWWTGIVLLAADD244 pKa = 4.13ALGLGEE250 pKa = 4.1RR251 pKa = 11.84WLRR254 pKa = 11.84LALSHH259 pKa = 6.94RR260 pKa = 11.84DD261 pKa = 2.94KK262 pKa = 11.15HH263 pKa = 5.82HH264 pKa = 7.0ASRR267 pKa = 11.84TAALQCQRR275 pKa = 11.84DD276 pKa = 3.56MLLRR280 pKa = 11.84QRR282 pKa = 11.84RR283 pKa = 11.84RR284 pKa = 11.84ARR286 pKa = 11.84RR287 pKa = 11.84LHH289 pKa = 5.93AVSEE293 pKa = 4.5GKK295 pKa = 9.93LQEE298 pKa = 4.03EE299 pKa = 5.07KK300 pKa = 10.52KK301 pKa = 10.37RR302 pKa = 11.84QSALVWNVEE311 pKa = 3.71ARR313 pKa = 11.84PFPSTHH319 pKa = 5.48QLIVLPPPVASAPPAVPSQPPEE341 pKa = 3.92YY342 pKa = 10.41SSVFPPVV349 pKa = 2.78
MM1 pKa = 7.61GNPRR5 pKa = 11.84SPLDD9 pKa = 3.11RR10 pKa = 11.84RR11 pKa = 11.84LRR13 pKa = 11.84FRR15 pKa = 11.84NVPFLPPRR23 pKa = 11.84IAASSTTTARR33 pKa = 11.84SLKK36 pKa = 10.05AKK38 pKa = 8.48TMEE41 pKa = 3.93MRR43 pKa = 11.84FTIAWMWFPSVLLILGLLTPPSNGCTVDD71 pKa = 2.93VGRR74 pKa = 11.84NVSIRR79 pKa = 11.84EE80 pKa = 3.74QCRR83 pKa = 11.84LRR85 pKa = 11.84NGATFSKK92 pKa = 10.98GDD94 pKa = 3.37IEE96 pKa = 5.57GNFSGPVVVEE106 pKa = 4.44LDD108 pKa = 3.82YY109 pKa = 11.77EE110 pKa = 4.91DD111 pKa = 4.89IDD113 pKa = 3.73ITGEE117 pKa = 3.9RR118 pKa = 11.84QRR120 pKa = 11.84LRR122 pKa = 11.84FHH124 pKa = 7.13LSGLGCPTKK133 pKa = 10.92EE134 pKa = 4.84NIRR137 pKa = 11.84KK138 pKa = 10.05DD139 pKa = 3.53NEE141 pKa = 4.06SDD143 pKa = 3.66VNGGIRR149 pKa = 11.84WALYY153 pKa = 9.78IQTGDD158 pKa = 2.97AKK160 pKa = 10.84YY161 pKa = 10.53GIRR164 pKa = 11.84NQHH167 pKa = 5.28LSIRR171 pKa = 11.84LMYY174 pKa = 10.14PGEE177 pKa = 4.51KK178 pKa = 8.85NTQQLLGSDD187 pKa = 4.87FSCEE191 pKa = 3.4RR192 pKa = 11.84HH193 pKa = 5.67RR194 pKa = 11.84RR195 pKa = 11.84PSTPLGKK202 pKa = 9.93NAEE205 pKa = 4.32VPPATRR211 pKa = 11.84TSSTYY216 pKa = 10.31GVLSAFVVWIGSGLNIIWWTGIVLLAADD244 pKa = 4.13ALGLGEE250 pKa = 4.1RR251 pKa = 11.84WLRR254 pKa = 11.84LALSHH259 pKa = 6.94RR260 pKa = 11.84DD261 pKa = 2.94KK262 pKa = 11.15HH263 pKa = 5.82HH264 pKa = 7.0ASRR267 pKa = 11.84TAALQCQRR275 pKa = 11.84DD276 pKa = 3.56MLLRR280 pKa = 11.84QRR282 pKa = 11.84RR283 pKa = 11.84RR284 pKa = 11.84ARR286 pKa = 11.84RR287 pKa = 11.84LHH289 pKa = 5.93AVSEE293 pKa = 4.5GKK295 pKa = 9.93LQEE298 pKa = 4.03EE299 pKa = 5.07KK300 pKa = 10.52KK301 pKa = 10.37RR302 pKa = 11.84QSALVWNVEE311 pKa = 3.71ARR313 pKa = 11.84PFPSTHH319 pKa = 5.48QLIVLPPPVASAPPAVPSQPPEE341 pKa = 3.92YY342 pKa = 10.41SSVFPPVV349 pKa = 2.78
Molecular weight: 39.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
66935 |
47 |
2241 |
371.9 |
41.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.982 ± 0.202 | 2.461 ± 0.107 |
4.968 ± 0.127 | 5.396 ± 0.146 |
3.727 ± 0.111 | 6.233 ± 0.221 |
3.172 ± 0.092 | 3.435 ± 0.129 |
2.903 ± 0.098 | 10.352 ± 0.213 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.056 ± 0.071 | 3.266 ± 0.131 |
5.922 ± 0.204 | 3.665 ± 0.2 |
7.74 ± 0.187 | 7.772 ± 0.23 |
6.784 ± 0.221 | 7.498 ± 0.155 |
1.43 ± 0.083 | 3.239 ± 0.108 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
