
Microbacterium sp. ZXX196
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium; unclassified Microbacterium
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
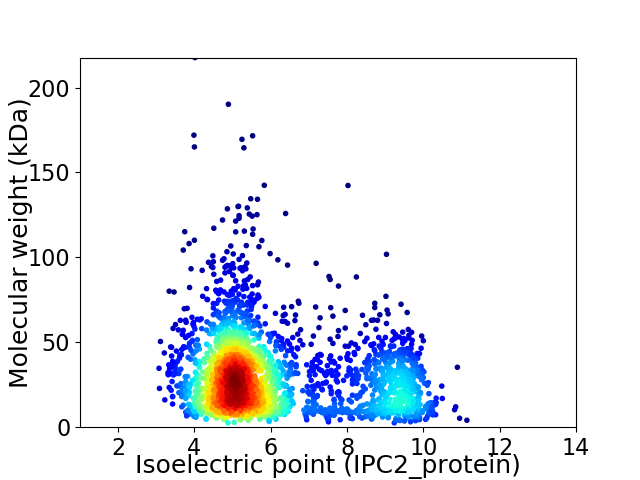
Virtual 2D-PAGE plot for 2672 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6I3LAB8|A0A6I3LAB8_9MICO 50S ribosomal protein L36 OS=Microbacterium sp. ZXX196 OX=2609291 GN=rpmJ PE=3 SV=1
MM1 pKa = 8.12RR2 pKa = 11.84LTTPLAAAALAGALVLTSCSSSQNPGPDD30 pKa = 3.11EE31 pKa = 4.74GGVAQADD38 pKa = 3.39ATIYY42 pKa = 10.6VGSLYY47 pKa = 10.49EE48 pKa = 4.16PQNLDD53 pKa = 3.24NTSGGGQGVTEE64 pKa = 4.27ALTGNVYY71 pKa = 10.62EE72 pKa = 4.32GLYY75 pKa = 10.48RR76 pKa = 11.84LADD79 pKa = 3.72SGEE82 pKa = 4.11VEE84 pKa = 4.36PLLAAAHH91 pKa = 5.84EE92 pKa = 4.26VSEE95 pKa = 5.53DD96 pKa = 3.01GLTYY100 pKa = 10.66TFTLQDD106 pKa = 3.44GVSFASGKK114 pKa = 8.06TLTADD119 pKa = 3.25DD120 pKa = 3.84VVYY123 pKa = 10.88SIEE126 pKa = 4.73KK127 pKa = 10.44VIADD131 pKa = 4.21DD132 pKa = 3.72SQSARR137 pKa = 11.84KK138 pKa = 8.82SQLEE142 pKa = 4.4VIDD145 pKa = 4.13SVAAVDD151 pKa = 3.91DD152 pKa = 3.5QTVRR156 pKa = 11.84VTLSEE161 pKa = 3.94RR162 pKa = 11.84SISFLYY168 pKa = 9.67NLSYY172 pKa = 11.06VWIVNSAFEE181 pKa = 4.46GNRR184 pKa = 11.84SSEE187 pKa = 4.14ADD189 pKa = 3.1GTGPYY194 pKa = 10.56VLDD197 pKa = 2.97TWTRR201 pKa = 11.84GSALSLVRR209 pKa = 11.84NDD211 pKa = 4.95DD212 pKa = 3.58YY213 pKa = 11.25WGEE216 pKa = 3.92APANGGVVFTYY227 pKa = 10.58FSDD230 pKa = 3.58ATSLNNALLTGAVDD244 pKa = 3.79VVTSQQSPDD253 pKa = 2.99GLAQFEE259 pKa = 4.38QTDD262 pKa = 4.13DD263 pKa = 3.66FTITDD268 pKa = 3.87GNSTTKK274 pKa = 10.89LLLAFNDD281 pKa = 3.49AAAPFDD287 pKa = 4.02DD288 pKa = 3.58VRR290 pKa = 11.84VRR292 pKa = 11.84QAVSAAIDD300 pKa = 3.73DD301 pKa = 4.26EE302 pKa = 4.54KK303 pKa = 11.24LLEE306 pKa = 4.52SVWGDD311 pKa = 3.37YY312 pKa = 8.14GTLIGSMVPPTDD324 pKa = 2.97PWFEE328 pKa = 4.66DD329 pKa = 3.47LTDD332 pKa = 4.81LDD334 pKa = 4.35TYY336 pKa = 11.57DD337 pKa = 4.25PDD339 pKa = 3.78RR340 pKa = 11.84AADD343 pKa = 3.99LLAEE347 pKa = 4.72AGYY350 pKa = 11.07EE351 pKa = 4.09DD352 pKa = 5.53GFTFTLDD359 pKa = 3.3TPNYY363 pKa = 9.21DD364 pKa = 3.72PHH366 pKa = 6.24PTAATFIQSQLAEE379 pKa = 3.88VGITVEE385 pKa = 4.26INTITSDD392 pKa = 2.67EE393 pKa = 4.85WYY395 pKa = 9.44TKK397 pKa = 10.26VYY399 pKa = 9.93QNRR402 pKa = 11.84DD403 pKa = 3.31FEE405 pKa = 4.61ATMQEE410 pKa = 4.31HH411 pKa = 6.41VNDD414 pKa = 5.12RR415 pKa = 11.84DD416 pKa = 3.88LVWYY420 pKa = 8.66GNPDD424 pKa = 4.11FYY426 pKa = 10.72WGYY429 pKa = 11.4DD430 pKa = 3.3NAEE433 pKa = 3.99VQEE436 pKa = 4.23WVAQSEE442 pKa = 4.69LVSSEE447 pKa = 4.3AEE449 pKa = 3.82QTEE452 pKa = 4.19LLKK455 pKa = 11.08LVARR459 pKa = 11.84TTAEE463 pKa = 3.86EE464 pKa = 4.29AASEE468 pKa = 3.99WLYY471 pKa = 10.8LYY473 pKa = 9.17PQIVVASSALSGYY486 pKa = 8.67PVNGLNSQFFAYY498 pKa = 10.2DD499 pKa = 3.19IEE501 pKa = 4.48KK502 pKa = 10.66AAA504 pKa = 4.43
MM1 pKa = 8.12RR2 pKa = 11.84LTTPLAAAALAGALVLTSCSSSQNPGPDD30 pKa = 3.11EE31 pKa = 4.74GGVAQADD38 pKa = 3.39ATIYY42 pKa = 10.6VGSLYY47 pKa = 10.49EE48 pKa = 4.16PQNLDD53 pKa = 3.24NTSGGGQGVTEE64 pKa = 4.27ALTGNVYY71 pKa = 10.62EE72 pKa = 4.32GLYY75 pKa = 10.48RR76 pKa = 11.84LADD79 pKa = 3.72SGEE82 pKa = 4.11VEE84 pKa = 4.36PLLAAAHH91 pKa = 5.84EE92 pKa = 4.26VSEE95 pKa = 5.53DD96 pKa = 3.01GLTYY100 pKa = 10.66TFTLQDD106 pKa = 3.44GVSFASGKK114 pKa = 8.06TLTADD119 pKa = 3.25DD120 pKa = 3.84VVYY123 pKa = 10.88SIEE126 pKa = 4.73KK127 pKa = 10.44VIADD131 pKa = 4.21DD132 pKa = 3.72SQSARR137 pKa = 11.84KK138 pKa = 8.82SQLEE142 pKa = 4.4VIDD145 pKa = 4.13SVAAVDD151 pKa = 3.91DD152 pKa = 3.5QTVRR156 pKa = 11.84VTLSEE161 pKa = 3.94RR162 pKa = 11.84SISFLYY168 pKa = 9.67NLSYY172 pKa = 11.06VWIVNSAFEE181 pKa = 4.46GNRR184 pKa = 11.84SSEE187 pKa = 4.14ADD189 pKa = 3.1GTGPYY194 pKa = 10.56VLDD197 pKa = 2.97TWTRR201 pKa = 11.84GSALSLVRR209 pKa = 11.84NDD211 pKa = 4.95DD212 pKa = 3.58YY213 pKa = 11.25WGEE216 pKa = 3.92APANGGVVFTYY227 pKa = 10.58FSDD230 pKa = 3.58ATSLNNALLTGAVDD244 pKa = 3.79VVTSQQSPDD253 pKa = 2.99GLAQFEE259 pKa = 4.38QTDD262 pKa = 4.13DD263 pKa = 3.66FTITDD268 pKa = 3.87GNSTTKK274 pKa = 10.89LLLAFNDD281 pKa = 3.49AAAPFDD287 pKa = 4.02DD288 pKa = 3.58VRR290 pKa = 11.84VRR292 pKa = 11.84QAVSAAIDD300 pKa = 3.73DD301 pKa = 4.26EE302 pKa = 4.54KK303 pKa = 11.24LLEE306 pKa = 4.52SVWGDD311 pKa = 3.37YY312 pKa = 8.14GTLIGSMVPPTDD324 pKa = 2.97PWFEE328 pKa = 4.66DD329 pKa = 3.47LTDD332 pKa = 4.81LDD334 pKa = 4.35TYY336 pKa = 11.57DD337 pKa = 4.25PDD339 pKa = 3.78RR340 pKa = 11.84AADD343 pKa = 3.99LLAEE347 pKa = 4.72AGYY350 pKa = 11.07EE351 pKa = 4.09DD352 pKa = 5.53GFTFTLDD359 pKa = 3.3TPNYY363 pKa = 9.21DD364 pKa = 3.72PHH366 pKa = 6.24PTAATFIQSQLAEE379 pKa = 3.88VGITVEE385 pKa = 4.26INTITSDD392 pKa = 2.67EE393 pKa = 4.85WYY395 pKa = 9.44TKK397 pKa = 10.26VYY399 pKa = 9.93QNRR402 pKa = 11.84DD403 pKa = 3.31FEE405 pKa = 4.61ATMQEE410 pKa = 4.31HH411 pKa = 6.41VNDD414 pKa = 5.12RR415 pKa = 11.84DD416 pKa = 3.88LVWYY420 pKa = 8.66GNPDD424 pKa = 4.11FYY426 pKa = 10.72WGYY429 pKa = 11.4DD430 pKa = 3.3NAEE433 pKa = 3.99VQEE436 pKa = 4.23WVAQSEE442 pKa = 4.69LVSSEE447 pKa = 4.3AEE449 pKa = 3.82QTEE452 pKa = 4.19LLKK455 pKa = 11.08LVARR459 pKa = 11.84TTAEE463 pKa = 3.86EE464 pKa = 4.29AASEE468 pKa = 3.99WLYY471 pKa = 10.8LYY473 pKa = 9.17PQIVVASSALSGYY486 pKa = 8.67PVNGLNSQFFAYY498 pKa = 10.2DD499 pKa = 3.19IEE501 pKa = 4.48KK502 pKa = 10.66AAA504 pKa = 4.43
Molecular weight: 54.6 kDa
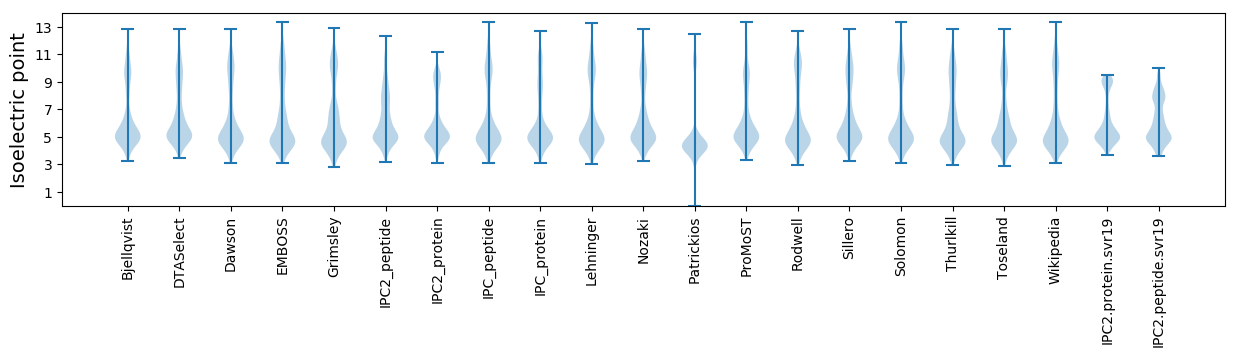
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6I3LDZ8|A0A6I3LDZ8_9MICO DUF368 domain-containing protein OS=Microbacterium sp. ZXX196 OX=2609291 GN=GJQ66_03800 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
793030 |
22 |
2073 |
296.8 |
31.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.316 ± 0.088 | 0.503 ± 0.012 |
6.326 ± 0.05 | 6.063 ± 0.044 |
3.136 ± 0.026 | 9.104 ± 0.045 |
1.928 ± 0.022 | 4.435 ± 0.033 |
1.95 ± 0.032 | 9.708 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.832 ± 0.021 | 1.847 ± 0.025 |
5.282 ± 0.035 | 2.677 ± 0.027 |
7.662 ± 0.067 | 5.193 ± 0.035 |
5.712 ± 0.03 | 8.897 ± 0.042 |
1.472 ± 0.022 | 1.958 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
