
Arthrobacter agilis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Arthrobacter
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
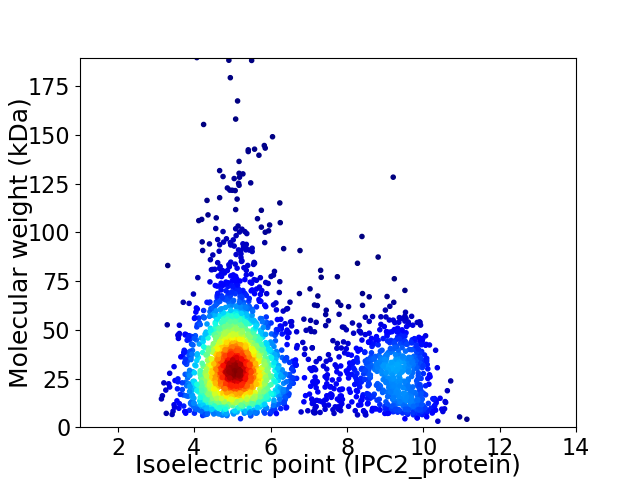
Virtual 2D-PAGE plot for 2887 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
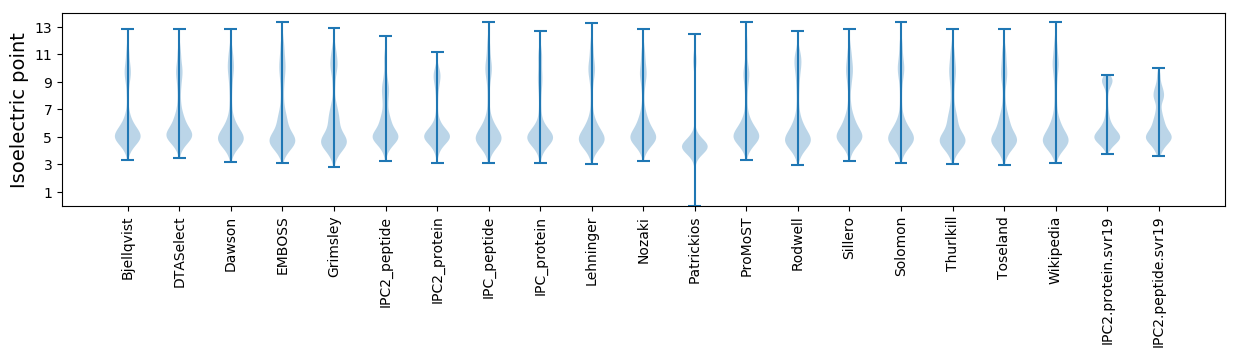
* You can choose from 21 different methods for calculating isoelectric point
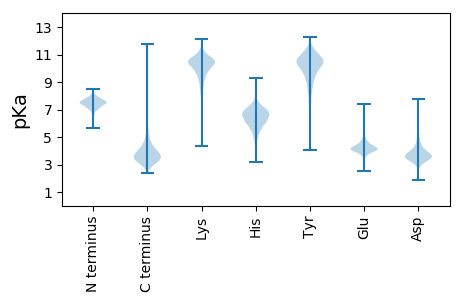
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y3LYI9|A0A1Y3LYI9_9MICC Cholesterol dehydrogenase OS=Arthrobacter agilis OX=37921 GN=B8W74_12855 PE=4 SV=1
MM1 pKa = 7.46KK2 pKa = 10.55AKK4 pKa = 10.05FLASAGIVAATAVALTGCSGGNGGGDD30 pKa = 3.31GGGSEE35 pKa = 5.13AASCTNTIVNAEE47 pKa = 4.06APQVTVWAWYY57 pKa = 8.09PAFDD61 pKa = 4.2QVVDD65 pKa = 3.79NFNNANDD72 pKa = 4.22DD73 pKa = 4.28VQICWTNAGQGNDD86 pKa = 3.61EE87 pKa = 4.16YY88 pKa = 10.85TKK90 pKa = 10.57FSTSIEE96 pKa = 4.43SGSGAPDD103 pKa = 3.87VIQLEE108 pKa = 4.61SEE110 pKa = 4.4VLSSFTIRR118 pKa = 11.84DD119 pKa = 3.45ALVDD123 pKa = 3.76LTEE126 pKa = 4.2YY127 pKa = 10.55GANDD131 pKa = 3.37VEE133 pKa = 4.28GDD135 pKa = 3.81YY136 pKa = 11.77AEE138 pKa = 5.0GAWEE142 pKa = 4.32DD143 pKa = 3.96ASSGDD148 pKa = 3.44AVYY151 pKa = 10.35AIPVDD156 pKa = 4.16GGPMGMLYY164 pKa = 10.34RR165 pKa = 11.84QDD167 pKa = 3.34IFDD170 pKa = 3.87AAGIEE175 pKa = 4.71VPTTWEE181 pKa = 3.85EE182 pKa = 3.88FEE184 pKa = 3.97AAARR188 pKa = 11.84TLKK191 pKa = 10.89DD192 pKa = 3.37SGDD195 pKa = 3.68GAVIADD201 pKa = 4.62FPTNGRR207 pKa = 11.84AFNQALFAQAGSEE220 pKa = 4.01PFVYY224 pKa = 10.49DD225 pKa = 3.13SANPQEE231 pKa = 4.57IGITVNDD238 pKa = 4.18DD239 pKa = 3.29GAKK242 pKa = 10.08KK243 pKa = 10.32VLEE246 pKa = 4.13YY247 pKa = 10.71WNGLVEE253 pKa = 5.45DD254 pKa = 4.77GLVATDD260 pKa = 4.26DD261 pKa = 4.86AFTADD266 pKa = 4.2YY267 pKa = 7.76NTKK270 pKa = 10.53LVDD273 pKa = 3.63GSYY276 pKa = 11.06AVYY279 pKa = 10.1LAAAWGPGYY288 pKa = 10.47LQGLSDD294 pKa = 4.5SDD296 pKa = 4.17PDD298 pKa = 4.22AVWRR302 pKa = 11.84AAPLPLWDD310 pKa = 4.08EE311 pKa = 4.45ANPVQVNWGGSTFAVTEE328 pKa = 3.77QAEE331 pKa = 4.47NKK333 pKa = 9.46EE334 pKa = 3.84LAARR338 pKa = 11.84VAMEE342 pKa = 3.78IFGNEE347 pKa = 3.19EE348 pKa = 3.76RR349 pKa = 11.84MNLGVDD355 pKa = 3.8EE356 pKa = 4.91GALFPSYY363 pKa = 10.88LPVLEE368 pKa = 4.55SPEE371 pKa = 4.19FEE373 pKa = 3.96NKK375 pKa = 9.72EE376 pKa = 3.87YY377 pKa = 10.97EE378 pKa = 4.34FFGGQQINKK387 pKa = 9.99DD388 pKa = 3.74VFLDD392 pKa = 3.84AAAGYY397 pKa = 9.57EE398 pKa = 4.19GATFSPFQNYY408 pKa = 10.17AYY410 pKa = 10.26DD411 pKa = 3.58QLTEE415 pKa = 3.82QIYY418 pKa = 11.32AMVQGEE424 pKa = 4.19KK425 pKa = 10.71DD426 pKa = 3.12ADD428 pKa = 3.6QALDD432 pKa = 4.0DD433 pKa = 4.68LQSSLEE439 pKa = 4.06QYY441 pKa = 9.34ATEE444 pKa = 3.81QGFTIKK450 pKa = 10.67
MM1 pKa = 7.46KK2 pKa = 10.55AKK4 pKa = 10.05FLASAGIVAATAVALTGCSGGNGGGDD30 pKa = 3.31GGGSEE35 pKa = 5.13AASCTNTIVNAEE47 pKa = 4.06APQVTVWAWYY57 pKa = 8.09PAFDD61 pKa = 4.2QVVDD65 pKa = 3.79NFNNANDD72 pKa = 4.22DD73 pKa = 4.28VQICWTNAGQGNDD86 pKa = 3.61EE87 pKa = 4.16YY88 pKa = 10.85TKK90 pKa = 10.57FSTSIEE96 pKa = 4.43SGSGAPDD103 pKa = 3.87VIQLEE108 pKa = 4.61SEE110 pKa = 4.4VLSSFTIRR118 pKa = 11.84DD119 pKa = 3.45ALVDD123 pKa = 3.76LTEE126 pKa = 4.2YY127 pKa = 10.55GANDD131 pKa = 3.37VEE133 pKa = 4.28GDD135 pKa = 3.81YY136 pKa = 11.77AEE138 pKa = 5.0GAWEE142 pKa = 4.32DD143 pKa = 3.96ASSGDD148 pKa = 3.44AVYY151 pKa = 10.35AIPVDD156 pKa = 4.16GGPMGMLYY164 pKa = 10.34RR165 pKa = 11.84QDD167 pKa = 3.34IFDD170 pKa = 3.87AAGIEE175 pKa = 4.71VPTTWEE181 pKa = 3.85EE182 pKa = 3.88FEE184 pKa = 3.97AAARR188 pKa = 11.84TLKK191 pKa = 10.89DD192 pKa = 3.37SGDD195 pKa = 3.68GAVIADD201 pKa = 4.62FPTNGRR207 pKa = 11.84AFNQALFAQAGSEE220 pKa = 4.01PFVYY224 pKa = 10.49DD225 pKa = 3.13SANPQEE231 pKa = 4.57IGITVNDD238 pKa = 4.18DD239 pKa = 3.29GAKK242 pKa = 10.08KK243 pKa = 10.32VLEE246 pKa = 4.13YY247 pKa = 10.71WNGLVEE253 pKa = 5.45DD254 pKa = 4.77GLVATDD260 pKa = 4.26DD261 pKa = 4.86AFTADD266 pKa = 4.2YY267 pKa = 7.76NTKK270 pKa = 10.53LVDD273 pKa = 3.63GSYY276 pKa = 11.06AVYY279 pKa = 10.1LAAAWGPGYY288 pKa = 10.47LQGLSDD294 pKa = 4.5SDD296 pKa = 4.17PDD298 pKa = 4.22AVWRR302 pKa = 11.84AAPLPLWDD310 pKa = 4.08EE311 pKa = 4.45ANPVQVNWGGSTFAVTEE328 pKa = 3.77QAEE331 pKa = 4.47NKK333 pKa = 9.46EE334 pKa = 3.84LAARR338 pKa = 11.84VAMEE342 pKa = 3.78IFGNEE347 pKa = 3.19EE348 pKa = 3.76RR349 pKa = 11.84MNLGVDD355 pKa = 3.8EE356 pKa = 4.91GALFPSYY363 pKa = 10.88LPVLEE368 pKa = 4.55SPEE371 pKa = 4.19FEE373 pKa = 3.96NKK375 pKa = 9.72EE376 pKa = 3.87YY377 pKa = 10.97EE378 pKa = 4.34FFGGQQINKK387 pKa = 9.99DD388 pKa = 3.74VFLDD392 pKa = 3.84AAAGYY397 pKa = 9.57EE398 pKa = 4.19GATFSPFQNYY408 pKa = 10.17AYY410 pKa = 10.26DD411 pKa = 3.58QLTEE415 pKa = 3.82QIYY418 pKa = 11.32AMVQGEE424 pKa = 4.19KK425 pKa = 10.71DD426 pKa = 3.12ADD428 pKa = 3.6QALDD432 pKa = 4.0DD433 pKa = 4.68LQSSLEE439 pKa = 4.06QYY441 pKa = 9.34ATEE444 pKa = 3.81QGFTIKK450 pKa = 10.67
Molecular weight: 48.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y3LV39|A0A1Y3LV39_9MICC Transcriptional regulator MalT OS=Arthrobacter agilis OX=37921 GN=B8W74_13310 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
945779 |
26 |
1816 |
327.6 |
35.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.011 ± 0.061 | 0.571 ± 0.011 |
6.185 ± 0.037 | 5.64 ± 0.041 |
3.089 ± 0.027 | 9.246 ± 0.042 |
2.036 ± 0.022 | 4.193 ± 0.034 |
2.032 ± 0.033 | 10.329 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.836 ± 0.018 | 2.065 ± 0.024 |
5.472 ± 0.034 | 2.964 ± 0.022 |
7.2 ± 0.053 | 5.951 ± 0.033 |
6.166 ± 0.033 | 8.673 ± 0.042 |
1.344 ± 0.018 | 1.995 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
